2. 南京林业大学, 南京 210037;
3. 玉林市林业科学研究所, 广西 容县 537501;
4. 广西壮族自治区林业科学研究院, 广西优良用材林资源培育重点实验室, 中南速生材繁育国家林业局重点实验室, 南宁 530002
2. Nanjing Forestry University, Nanjing 210037, China;
3. Yulin Forestry Research Institute, Rongxian 537501, Guangxi, China;
4. Guangxi Key Laboratory of Superior Timber Trees Resource Cultivation, Key Laboratory of Central South Fast-growing Timber Cultivation of Forestry Ministry of China, Gugangxi Forestry Research Institute, Nanning 530002, China
根据联合国粮农组织2020年的数据,全球森林面积占土地总面积的比例已经从2000年的31.9%降至31.2%,木材资源短缺已经成为国际社会公认的发展障碍问题之一。随着天然林资源的减少、经济水平的快速发展以及人均住宅面积的不断增加,我国木材资源的供需矛盾也随之加剧。当前我国木材对外依存度已超过50%,亟需培育更多实木用材缓解木材供需矛盾[1]。桉树人工林是世界三大速生人工林之一,桉属(Eucalyptus)一些树种具有轮伐期短、生物量大、干形好等特点,其木材产品经济价值高,可用作纸浆原料、家具和工业用材等[2]。我国桉树人工林及其产业发展非常迅速,桉树木材年产量超过4.0×107m3 [3]。目前我国桉树人工林主要以巨桉(E. grandis)、尾叶桉(E. urophylla)和细叶桉(E. tereticornis)等速生树种为主,而珍贵用材人工林面积占比少,大花序桉(E. cloeziana),市场俗称“澳洲大花梨”[4],是我国南方仅有的几个速生珍贵用材树种之一。大花序桉木材密度远高于短轮伐期的其它速生桉,其树干通直,结构均匀,生长速度快,适应性好,在我国华南地区广泛栽培,市场潜力巨大[1, 5]。
林木成熟木质部一般由边材、过渡区和心材构成,心材是珍贵树种的主要用材部分,是体现珍贵树种木材质量与价值的主体。维管形成层细胞的不断分裂、分化、细胞扩增、次生壁沉积、木质化,最后细胞程序性死亡使外周的边材逐渐转变为心材[6]。心边材转化过程可以反映林木的径向变化[7–8],同时木质部次生细胞壁生长会影响木材基本密度和力学强度等物理性质[9]。木材的物理性质既影响木材的力学强度,又决定着木材的产量和品质,已经作为林木遗传改良的重要育种目标[10–12],已有研究者在桉树中定位到影响木材物理性质的部分功能基因[13–14]。木材形成涉及众多生物学过程,受多层次、多基因的协同调控,也受环境影响,是一个复杂的发育过程[15–16]。遗传变异是木材形成的一个重要驱动因子[17–20]。挖掘边心材形成相关功能基因,可为培育心材比例高、材性优质、材色佳的优良种质资源,促进我国珍贵用材树种建设,以缓解珍贵用材资源匮乏以及市场供不应求的局面。桉树心边材变化的遗传规律与木材形成机制还不明确,挖掘边心材形成相关功能基因有助于解析桉树木材形成的遗传机制。
随着二代测序技术的高速发展,基于重测序开发单核苷酸多态性(single nucleotide polymorphism, SNP)位点已越来越普遍,目前广泛应用于桉树候选功能基因挖掘研究[21–23]。Bulk DNA测序可以快速筛选等位基因频率差异显著的SNP位点,进而挖掘复杂性状关联的候选功能基因[24–25],如毛果杨(Populus trichocarpa)生长[26]、鹰嘴豆(Cicer arietinum)荚果数[27]和胡麻(Linum usitatissimum)株高[28]等,但在林木材性方面研究应用报道较少。因此, 本研究利用广西玉林18 a生大花序桉种源家系试验林, 筛选心材差异显著的2个家系,各制作3株解析木,并采集胸径处初生木质部DNA,通过Bulk DNA测序筛选与心材变异相关候选基因。通过探究不同家系间边心材径向生长变化规律及挖掘相关候选功能基因,为解析大花序桉木材形成机制提供理论依据。
1 材料和方法 1.1 样木圆盘取样与处理试验材料来源于2004年5月种植于广西玉林市林业科学研究所(110°09′ E, 22°39′ N)的大花序桉种源/家系试验林。该试验林利用天然林按单株母树(间隔100 m以上)采集的种子,1株母树的子代即为1个半同胞家系,于2003年育苗,试验林抚育及施肥措施一致。筛选出心材比例差异显著的2个18 a生大花序桉家系(家系1: C4119,家系2: C4080)各3株制备解析木。树木伐倒前,在树干上分别标记东、北2个方位及胸高位置。伐倒后,测量树高及枝下高,并在树干基部0 m和胸高1.3 m处各取1个厚度5 cm的圆盘。胸高以上的木段每隔2 m截取1个圆盘,并编号,标出东、北方向,直至距树梢不足1 m处为止。通过对树干顶部的纵向解剖,测量心材消失处的高度。将圆盘表面刨光,使心材部分清晰可见,使用杭州万深LA-S植物图像分析系统对圆盘进行扫描并进行图像分析处理。分别测量圆盘4个方向的树干去皮半径(xylem radius, XR)、心材半径(heartwood radius, HR)和边材宽度(sapwood width, SW),将4个方向的平均值作为该圆盘的横截面去皮直径、去皮半径、心材半径和边材宽度。以单株树木不同圆盘上心材半径和边材宽度的中位数估算单株水平的心、边材量,以避免树木基部膨大及树干局部形变对平均值的影响。心材面积(heartwood area, HA)=以心材为半径圆的面积、边材面积(sapwood area, SA)=圆盘面积-心材面积、心材率(heartwood rate, HR)=心材占圆盘界面的面积比[29]。基本密度和木材力学性能采用GB/T 1927.5—2021无疵小试样木材物理力学性质试验方法[30]。
1.2 候选基因筛选及注释分析分别取各解析木胸径处初生木质部材料,用锡纸包裹后立刻用液氮冷冻保存,采用CTAB法提取基因组DNA,Qubit 3.0 (ThermoFisher, USA)精确定量。同一个家系的3个样品DNA等量混合后打断为300 bp片段,利用VAHTSTM Universal DNA Library Prep Kit for Illumina构建基因组文库,文库质检后委托公司进行测序。Bulk DNA测序数据使用Trim-momatic-0.36[31]过滤低质量序列后,利用samtools v 1.7[32]和bwa 0.9[33]比对到巨桉(v 2.0)参考基因组(https://phytozome.jgi.doe.gov/pz/portal.html), 通过VarScan2查找SNP[34]后采用VCFtools[35]过滤低质量位点,筛选两家系间基因频率差异在60%以上的SNP位点,并使用SnpEff v 4.3[36]对位点进行注释分析。以SNP位点上下游各1 000 bp为区间,利用巨桉基因组注释信息获取SNP点区域候选功能基因信息并进行GO[37–38]和KEGG[39]富集分析(http://kobas.cbi.pku.edu.cn)。
1.3 数据分析分析两家系间方位变异,心材半径与年轮随树高变化并拟合回归方程,解析心边材比例变化规律。对家系间心材半径和边材宽度的方位变异、心材比例进行方差分析(ANOVA, F检验)和两样本平均数检验(t检验),采用线性模型拟合心材半径与心材年轮个数随树高的回归方程,图形绘制用R 4.2.1软件完成。
2 结果和分析 2.1 边材宽度与心材半径的方位变异大花序桉家系2的边材宽度和心材半径在4个方向上均大于家系1 (图 1),2个家系的边材宽度和心材半径平均差值分别为0.7和5.5 cm,边材宽度均为东部最大,其中家系1为1.35 cm,家系2为2.15 cm。心材半径上两个家系间出现了不同的变化,家系1的西部和东部大于北部和南部,家系2的北部和东部大于西部和南部。总体来说,两个家系边材宽度和心材半径在4个方向上都无显著差异(P > 0.05)。
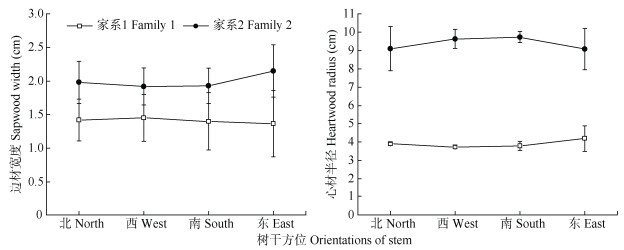
|
图 1 大花序桉边材宽度与心材半径的方位变异 Fig. 1 Orientations variation of sapwood width and heartwood radius in Eucalyptus cloeziana |
| 表 1 大花序桉解析木基本信息 Table 1 Timber data of Eucalyptus cloeziana |
家系1的3株解析木圆盘基部与顶部间的平均差值为7.4 cm,与家系2类似(7.6 cm),2个家系间心材半径的平均差值随高度变化也相似,都为6~7 cm。2个家系的心材年轮数在树木基部差距均为13~15,但随着高度增加,家系1的心材年轮逐渐小于家系2,在5.3 m后完全小于家系2,且顶部与基部年轮数平均差值都为11。两个家系间心材半径和心材年轮数随树高的增加而递减,与心材半径的下降趋势相似(图 2)。一次函数很好拟合了心材半径和心材年轮数与树高的关系,其中家系1与家系2在心材半径和心材年轮数上分别解释了89%和95%,97%和96%的变异。心材半径上家系1的下降速率略小于家系2,而心材年轮个数上家系1的下降速率大于家系2 (表 2)。
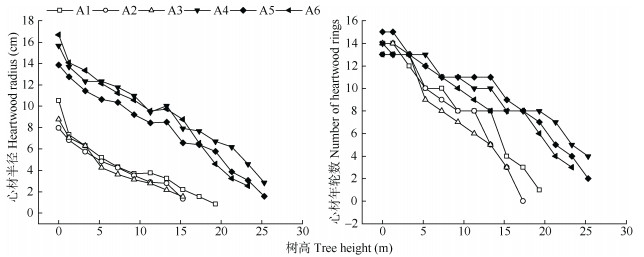
|
图 2 大花序桉心材半径和心材年轮个数随树高的变异。A1~A6见表 1. Fig. 2 Variation of heartwood radius and rings number with tree height in Eucalyptus cloeziana. A1-A6 see Table 1. |
| 表 2 心材半径与年轮数与树高的回归方程 Table 2 Regression equation of tree height with radius and ring number of heartwood |
家系1和2的心材率随树高逐渐减小,边材率则逐渐增加。基部心材率最大,家系1和2非常接近,分别为74%和73%,顶部心材率差异较大,分别为17%和12%,而两家系的心材率和边材率随树高变化趋势相似。两个家系间的心材率和边材率出现的交汇点高度不同,其中家系1约为16 m,位于整体树高4/5处,而家系2为23 m,位于整体树高的9/10处。另外家系2在0~13.3 m处的心材率维持在70%左右,而家系1没有稳定的高心材率区间(图 3)。

|
图 3 大花序桉家系1与家系2的心材率和边材率 Fig. 3 Heartwood rate and sapwood rate of two Eucalyptus cloeziana families |
家系1和2木材的基本密度分别为0.80~0.82和0.75~0.78 g/cm3,且2个家系间差异显著,平均差值为0.048 g/cm3 (表 3)。基本密度与顺纹抗拉强度和弦面硬度呈正相关外,其他性状基本上为负相关, 其中与心材半径为显著负相关,皮尔逊相关系数–0.875,与树高呈极显著负相关。树高也和心材半径、弦面硬度、抗弯弹性模量之间差异显著。基本密度与弦面硬度呈显著正相关,皮尔逊相关系数为0.893。力学性质相关性分析表明,顺纹抗拉强度与胸径、心材半径和横截面半径呈显著负相关,皮尔逊系数分别为–0.921、–0.910和–0.913。弦面硬度与心材半径和横截面半径呈显著负相关,皮尔逊相关系数分别为–0.841和–0.824。
| 表 3 大花序桉木材基本密度相关性分析 Table 3 Correlation analysis of wood basic density in Eucalyptus cloezia |
通过VarScan2共计开发了16 941 942个SNP位点,其中两家系间等位基因频率差异60%以上的位点为647 873个,手动过滤低等位基因频率SNP位点后共筛选出1842个SNP位点。SNPeff注释分析表明,位于基因间的SNP位点最多,占比55.8%,其次为上游区域,占比18.3%,下游区域、外显子和内含子占比分别为16.3%、5.1%和4.4% (图 4)。

|
图 4 SNP位点分布图 Fig. 4 SNP locus distribution |
基因组滑窗分析以10K为指标进行SNP位点区间候选功能基因筛选。通过GO富集分析共注释262个terms (图 5: A),其中生物过程包含145个terms,主要和蛋白质代谢过程、蛋白质修饰以及蛋白质磷酸化相关;分子功能共富集到91个terms上,主要和离子结合、催化活性和有机环与杂环化合物相关;细胞组分富集到26个terms都和生物膜相关。KEGG富集分析发现有关生长激素合成相关的10条代谢途径(图 5: B)。
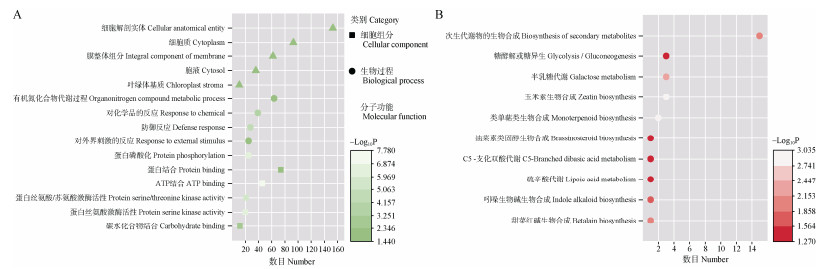
|
图 5 大花序桉心材变异相关候选基因的GO (A)和KEGG (B)富集分析 Fig. 5 GO enrichment (A) and KEGG enrichment analysis (B) of candidate genes related to heartwood variation in Eucalyptus cloezia |
早期研究表明, 树高、胸径以及树冠的相对高度和边材宽度呈正相关[40]。本研究中家系2的边材宽度与心材半径都大于家系1,这可能是由于家系2的平均树高和胸径都大于家系1。但本研究中边材宽度和心材宽度的方位变异不一致,这在油松中也有类似的研究结果[41],侧枝生物量、养分以及土壤水热微环境变化影响了树木生理过程,因此也会体现在林木边心材的方位变异上[42–44]。家系1与家系2心材半径随树高变化的趋势相似,都是随树高逐渐下降,心材半径的下降速率均为0.4,这与罗佳等[1]对不同林龄大花序桉的线性变化研究一致, 17 a生的心材半径下降率达到了0.29,而5、29和35 a生则维持在0.4左右[1]。本研究中18 a生大花序桉两个家系心边材方位变异上有差异,心材年轮数下降速率变化分别为0.64和0.36,两个家系的心材率也不同,且心材率与边材率出现的交汇位点也不同,出现此差异可能是树木内部的发育变化受到激素水平的调控和水分等因素的影响[45]。
热带树种的木材基本密度大多和生长速率、胸径等呈负相关[46–47],在本研究中,树高和横截面半径与木材密度呈负相关,同时心材半径也和基本密度呈显著负相关,而与边材宽度相关性低。木材密度低可能使其具有更大纤维直径和孔隙来储存水分[48],而含水量变化对木材直径变化与木质部细胞的发育影响较大[6, 49]。本研究表明家系1的平均基本密度大于家系2,家系2的树高、胸径和心材半径等生长指标均大于家系1,两个家系间基本密度的差异可能导致其对含水率和水分利用的不同, 进而间接影响了心边材生长差异变化。
富集分析表明,GO terms主要和植物细胞分裂、植物细胞膜、植物蛋白激酶以及核苷酸代谢有关, 杨树和桉树的木材形成过程还伴随核苷酸和碳水化合物的代谢产物变化[50–51]。本研究中注释到的4个细胞组分GO terms都富集到生物膜功能上,相关表达基因与细胞色素P450基因家族相关,该家族主要是调控合成植物次生代谢物的关键酶[52],与合成植物萜类化合物、植物激素、植物生长和非生物胁迫等功能高度相关[53]。木质部细胞类型、结构和化学组成(木质素和纤维素含量等)决定了木材品质[26], 一些P450家族和木质素合成中间产物以及酶相关[54], 如CYP71和CYP706家族催化植物类黄酮物质[55]及CYP73家族催化肉桂酸合成等[56],基因Eucgr. D00210和Eucgr.D00213与拟南芥中催化类黄酮合成的类似酶CYP706A4同源[57]。同时也发现和生长素合成相关的基因,如基因Eucgr. B01522与在拟南芥中合成生长素有关CYP79B2同源[58],上述酶都和调控植物激素合成相关,而植物激素是影响木质部细胞和木质部直径的直接因素[6]。这些基因可能参与大花序桉植物激素或者其他次生代谢物的合成,进而影响木质部的生长[52],但在大花序桉中的具体功能还需进一步验证。
另外富集分析表明,蛋白激酶基因主要与丝/苏氨酸蛋白激酶和受体蛋白激酶同源,这些酶在植物抗逆性、生长发育和信号传导等方面起重要调控作用[58]。次生细胞壁是木材主要组成部分,本研究发现基因Eucgr.F04299和Eucgr.E00980与细胞壁相关激酶(wall-associated kinase, WAK)同源,该酶参与调控拟南芥次生细胞壁的合成[59]。另外,定位的1个等位基因频率差异显著的SNP位点位于基因Eucgr.K01600和Eucgr.K01601之间,这2个基因和油菜素内酯受体激酶相关,对杨树、番茄(Lycopersicon esculentum)和水芹(Lepidium sativum)等植物的研究表明油菜素内酯参与调控植物细胞壁木质化[60–62],也参与木材边材向心材转化[6]。心边材的形成涉及植物代谢物组成、细胞分化和信号转导等众多生物过程,本研究通过对心边材差异显著的2个18 a生大花序桉家系的解析木分析,结合Bulk DNA测序分析,初步挖掘了一些大花序桉生长发育和材性相关的候选功能基因。大部分候选功能基因主要和细胞分裂、生物膜、植物激素以及木质素合成相关,KEGG富集到和玉米素合成(Eucgr.F00346和Eucgr.F00342)以及木质素合成(Eucgr.F01418和Eucgr.H02573)相关的苯丙氨酸代谢途径,这为后期开展大花序桉心材形成机制研究以及珍贵用材树种的分子育种奠定基础。
| [1] |
LUO J, MA R K, WEI P L, et al. Variation on radial and axial of heartwood and sapwood in Eucalyptus cloeziana[J]. J Beijing For Univ, 2021, 43(4): 132-140. 罗佳, 马若克, 韦鹏练, 等. 大花序桉心边材的径向和轴向的变异[J]. 北京林业大学学报, 2021, 43(4): 132-140. DOI:10.12171/j.1000-1522.20210021 |
| [2] |
CHEN Y P, LYU J X, CHEN Z L. Status of development and wood utilization of Eucalyptus plantation in China[J]. China Wood-Based Panels, 2019, 26(12): 6-9. 陈勇平, 吕建雄, 陈志林. 我国桉树人工林发展概况及其利用现状[J]. 中国人造板, 2019, 26(12): 6-9. DOI:10.3969/j.issn.1673-5064.2019.12.002 |
| [3] |
XIE Y J. Scientific innovation leads to fast development of eucalypt research and industry in China[J]. Eucalypt Sci Technol, 2022, 39(1): 35-42. 谢耀坚. 科技创新引领中国桉树研究和产业迅猛发展[J]. 桉树科技, 2022, 39(1): 35-42. DOI:10.13987/j.cnki.askj.2022.01.006 |
| [4] |
QI S X. Eucalyptus in China[M]. Beijing: China Forestry Publishing House, 2002. 祁述雄. 中国桉树[M]. 北京: 中国林业出版社, 2002. |
| [5] |
LI C R, CHEN K, ZHOU X J. Research status and development trend of Eucalyptus cloeziana[J]. Eucalypt Sci Technol, 2012, 29(2): 40-46. 李昌荣, 陈奎, 周小金. 大花序桉研究现状与发展趋势[J]. 桉树科技, 2012, 29(2): 40-46. DOI:10.3969/j.issn.1674-3172.2012.02.008 |
| [6] |
RATHGEBER C B K, CUNY H E, FONTI P. Biological basis of tree-ring formation: A crash course[J]. Front Plant Sci, 2016, 7: 734. DOI:10.3389/fpls.2016.00734 |
| [7] |
ZHOU Y F, DONG L H, LI F R. Growth variation characteristics of heart sapwood and bark of Populus nigra×P. simonii[J]. J NE For Univ, 2022, 50(4): 89-94. 周翼飞, 董利虎, 李凤日. 迎春5号杨树心边材及树皮的生长变异特征[J]. 东北林业大学学报, 2022, 50(4): 89-94. DOI:10.3969/j.issn.1000-5382.2022.04.015 |
| [8] |
WANG X C, WANG C K, ZHANG Q Z, et al. Growth characteristics of heartwood and sapwood of the major tree species in northeastern China[J]. Sci Silv Sin, 2008, 44(5): 102-108. 王兴昌, 王传宽, 张全智, 等. 东北主要树种心材与边材的生长特征[J]. 林业科学, 2008, 44(5): 102-108. DOI:10.3321/j.issn:1001-7488.2008.05.020 |
| [9] |
SUN H Y, SU M L, LÜ J X, et al. Research progress on effect of microfibril angle and crystalline area in cell wall on wood physical and mechanical properties[J]. J NW Agric For Univ (Nat Sci), 2019, 47(5): 50-58. 孙海燕, 苏明垒, 吕建雄, 等. 细胞壁微纤丝角和结晶区对木材物理力学性能影响研究进展[J]. 西北农林科技大学学报(自然科学版), 2019, 47(5): 50-58. DOI:10.13207/j.cnki.jnwafu.2019.05.007 |
| [10] |
MCLEAN J P, MOORE J R, GARDINER B A, et al. Variation of radial wood properties from genetically improved Sitka spruce growing in the UK[J]. Forestry, 2016, 89(2): 109-116. DOI:10.1093/forestry/cpv035 |
| [11] |
RODRÍGUEZ-PÉREZ D, MOYA R, MURILLO O, et al. Variation and genetic control of the heartwood, sapwood, bark, wood color parameter, and physical and mechanical properties of Dipteryx panamensis in Costa Rica[J]. Forests, 2022, 13(1): 106. DOI:10.3390/f13010106 |
| [12] |
CHU D M, YAO T, ZHOU L, et al. Genetic variation analysis and comprehensive evaluation of wood property traits of 20-year-old Chinese fir clone[J]. Eur J For Res, 2022, 141(1): 59-69. DOI:10.1007/s10342-021-01426-4 |
| [13] |
THUMMA B R, NOLAN M F, EVANS R, et al. Polymorphisms in Cinnamoyl CoA reductase (CCR) are associated with variation in microfibril angle in Eucalyptus spp.[J]. Genetics, 2005, 171(3): 1257-1265. DOI:10.1534/genetics.105.042028 |
| [14] |
ZHOU C P, WANG L, WENG Q J, et al. Association of microsatellite markers with growth and wood mechanical traits in Eucalyptus cloeziana F. Muell. (Myrtaceae)[J]. Ind Crops Prod, 2020, 154: 112702. DOI:10.1016/j.indcrop.2020.112702 |
| [15] |
GION J M, CAROUCHÉ A, DEWEER S, et al. Comprehensive genetic dissection of wood properties in a widely-grown tropical tree: Eucalyptus[J]. BMC Genom, 2011, 12: 301. DOI:10.1186/1471-2164-12-301 |
| [16] |
FREEMAN J S, POTTS B M, DOWNES G M, et al. Stability of quantitative trait loci for growth and wood properties across multiple pedigrees and environments in Eucalyptus globulus[J]. New Phytol, 2013, 198(4): 1121-1134. DOI:10.1111/nph.12237 |
| [17] |
TARDIF J, FLANNIGAN M, BERGERON Y. An analysis of the daily radial activity of 7 boreal tree species, northwestern Quebec[J]. Environ Monit Assess, 2001, 67(1/2): 141-160. DOI:10.1023/a:1006430422061 |
| [18] |
YE Z H, ZHONG R Q. Molecular control of wood formation in trees[J]. J Exp Bot, 2015, 66(14): 4119-4131. DOI:10.1093/jxb/erv081 |
| [19] |
ZHANG J, XIE M, TUSKAN G A, et al. Recent advances in the transcriptional regulation of secondary cell wall biosynthesis in the woody plants[J]. Front Plant Sci, 2018, 9: 1535. DOI:10.3389/fpls.2018.01535 |
| [20] |
WANG Y H, ZHAO C J, ZHOU W W, et al. Diurnal variation in stem radial growth of Eucalyptus robusta and its response to environmental factors[J]. J Tianjin Norm Univ (Nat Sci), 2017, 37(6): 31-36. 王艺涵, 赵从举, 周雯雯, 等. 桉树树干径向生长日变化及其对环境因子的响应[J]. 天津师范大学学报(自然科学版), 2017, 37(6): 31-36. DOI:10.3969/j.issn.1671-1114.2017.06.007 |
| [21] |
MHOSWA L, O'NEILL M M, MPHAHLELE M M, et al. A genomewide association study for resistance to the insect pest Leptocybe invasa in Eucalyptus grandis reveals genomic regions and positional candidate defense genes[J]. Plant Cell Physiol, 2020, 61(7): 1285-1296. DOI:10.1093/pcp/pcaa057 |
| [22] |
DASGUPTA M G, BARI M P A, SHANMUGAVEL S, et al. Targeted re-sequencing and genome-wide association analysis for wood property traits in breeding population of Eucalyptus tereticornis×E. grandis[J]. Genomics, 2021, 113(6): 4276-4292. DOI:10.1016/j.ygeno.2021.11.013 |
| [23] |
ESTOPA R A, PALUDETO J G Z, MÜLLER B S F, et al. Genomic prediction of growth and wood quality traits in Eucalyptus benthamii using different genomic models and variable SNP genotyping density [J/OL]. New For, 2022. doi: 10.1007/s11056-022-09924-y.
|
| [24] |
MICHELMORE R W, PARAN I, KESSELI R V. Identification of markers linked to disease-resistance genes by bulked segregant analysis: A rapid method to detect markers in specific genomic regions by using segregating populations[J]. Proc Natl Acad Sci USA, 1991, 88(21): 9828-9832. DOI:10.1073/pnas.88.21.9828 |
| [25] |
TAKAGI H, ABE A, YOSHIDA K, et al. QTL-seq: Rapid mapping of quantitative trait loci in rice by whole genome resequencing of DNA from two bulked populations[J]. Plant J, 2013, 74(1): 174-183. DOI:10.1111/tpj.12105 |
| [26] |
DU Q Z, GONG C R, WANG Q S, et al. Genetic architecture of growth traits in Populus revealed by integrated quantitative trait locus (QTL) analysis and association studies[J]. New Phytol, 2016, 209(3): 1067-1082. DOI:10.1111/nph.13695 |
| [27] |
DAS S, SINGH M, SRIVASTAVA R, et al. mQTL-seq delineates functionally relevant candidate gene harbouring a major QTL regulating pod number in chickpea[J]. DNA Res, 2016, 23(1): 53-65. DOI:10.1093/dnares/dsv036 |
| [28] |
SONG X X, WANG L M, ZHANG J P, et al. QTL mapping and function analysis of candidate genes related to plant height in flax[J]. J Agric Sci Technol, 2020, 22(6): 26-32. 宋夏夏, 王利民, 张建平, 等. 胡麻株高QTL定位与候选基因功能分析[J]. 中国农业科技导报, 2020, 22(6): 26-32. DOI:10.13304/j.nykjdb.2020.0142 |
| [29] |
DENG L P, REN S H, LU J X, et al. Growth characteristics and variation of heartwood and sapwood of Catalpa bungei[J]. Chin J Wood Sci Technol, 2019, 33(4): 9-13. 邓丽萍, 任素红, 吕建雄, 等. 楸树心材与边材的生长特征及变异规律[J]. 木材工业, 2019, 33(4): 9-13. DOI:10.19455/j.mcgy.20190403 |
| [30] |
State Administration for Market Regulation, Standardization Administration. GB/T 1927.10—2021 Test methods for physical and mechanical properties of small clear wood specimens Part 10: Determination of modulus of elasticity in bending[S]. Beijing: Standards Press of China, 2021. 国家市场监督管理总局, 国家标准化管理委员会. GB/T 1927.10—2021无疵小试样木材物理力学性质试验方法第10部分: 抗弯弹性模量测定[S]. 北京: 中国标准出版社, 2021. |
| [31] |
BOLGER A M, LOHSE M, USADEL B. Trimmomatic: A flexible trimmer for Illumina sequence data[J]. Bioinformatics, 2014, 30(15): 2114-2120. DOI:10.1093/bioinformatics/btu170 |
| [32] |
LI H, HANDSAKER B, WYSOKER A, et al. The sequence alignment/map format and SAMtools[J]. Bioinformatics, 2009, 25(16): 2078-2079. DOI:10.1093/bioinformatics/btp352 |
| [33] |
LI H, DURBIN R. Fast and accurate long-read alignment with Burrows-Wheeler transform[J]. Bioinformatics, 2010, 26(5): 589-595. DOI:10.1093/bioinformatics/btp698 |
| [34] |
KOBOLDT D C, ZHANG Q Y, LARSON D E, et al. VarScan 2: Somatic mutation and copy number alteration discovery in cancer by exome sequencing[J]. Genome Res, 2012, 22(3): 568-576. DOI:10.1101/gr.129684.111 |
| [35] |
DANECEK P, AUTON A, ABECASIS G, et al. The variant call format and VCFtools[J]. Bioinformatics, 2011, 27(15): 2156-2158. DOI:10.1093/bioinformatics/btr330 |
| [36] |
CINGOLANI P, PLATTS A, WANG L L, et al. A program for annotating and predicting the effects of single nucleotide polymorphisms, SnpEff: SNPs in the genome of Drosophila melanogaster strain w1118, iso-2, iso-3[J]. Fly, 2012, 6(2): 80-92. DOI:10.4161/fly.19695 |
| [37] |
ASHBURNER M, BALL C A, BLAKE J A, et al. Gene Ontology: Tool for the unification of biology[J]. Nat Genet, 2000, 25(1): 25-29. DOI:10.1038/75556 |
| [38] |
The Gene Ontology Consortium. The Gene Ontology resource: Enriching a GOld mine[J]. Nucl Acids Res, 2021, 49(D1): D325-D334. DOI:10.1093/nar/gkaa1113 |
| [39] |
BU D C, LUO H T, HUO P P, et al. KOBAS-i: Intelligent prioritization and exploratory visualization of biological functions for gene enrichment analysis[J]. Nucl Acids Res, 2021, 49(W1): W317-W325. DOI:10.1093/nar/gkab447 |
| [40] |
MCDOWELL N, BARNARD H, BOND B J, et al. The relationship between tree height and leaf area: Sapwood area ratio[J]. Oecologia, 2002, 132(1): 12-20. DOI:10.1007/s00442-002-0904-x |
| [41] |
CHANG J G, LI X P, LIU S R, et al. Variations in amount and ring number of sapwood and heartwood of Pinus tabulaeformis[J]. Sci Silv Sin, 2009, 45(11): 76-82. 常建国, 李新平, 刘世荣, 等. 油松心边材量及年轮数的变异特征[J]. 林业科学, 2009, 45(11): 76-82. DOI:10.3321/j.issn:1001-7488.2009.11.013 |
| [42] |
LIU J L, WANG C K, ZHANG Q Z. Spatial variations in stem heart-wood and sapwood for Larix gmelinii trees with various differentiation classes[J]. Sci Silv Sin, 2014, 50(12): 114-121. 刘家霖, 王传宽, 张全智. 不同分化等级兴安落叶松树干心材和边材的空间变异[J]. 林业科学, 2014, 50(12): 114-121. DOI:10.11707/j.1001-7488.20141216 |
| [43] |
WULLSCHLEGER S D, KING A W. Radial variation in sap velocity as a function of stem diameter and sapwood thickness in yellow-poplar trees[J]. Tree Physiol, 2000, 20(8): 511-518. DOI:10.1093/treephys/20.8.511 |
| [44] |
MÖRLING T, VALINGER E. Effects of fertilization and thinning on heartwood area, sapwood area and growth in Scots pine[J]. Scand J For Res, 1999, 14(5): 462-469. DOI:10.1080/02827589950154168 |
| [45] |
PERROT-RECHENMANN C. Cellular responses to auxin: Division versus expansion[J]. Cold Spring Harb Perspect Biol, 2010, 2(5): a001446. DOI:10.1101/cshperspect.a001446 |
| [46] |
POORTER L, WRIGHT S J, PAZ H, et al. Are functional traits good predictors of demographic rates? Evidence from five neotropical forests[J]. Ecology, 2008, 89(7): 1908-1920. DOI:10.1890/07-0207.1 |
| [47] |
FRANCIS E J, MULLER-LANDAU H C, WRIGHT S J, et al. Quantifying the role of wood density in explaining interspecific variation in growth of tropical trees[J]. Glob Ecol Biogeogr, 2017, 26(10): 1078-1087. DOI:10.1111/geb.12604 |
| [48] |
WERDIN J, FLETCHER T D, RAYNER J P, et al. Biochar made from low density wood has greater plant available water than biochar made from high density wood[J]. Sci Total Environ, 2020, 705: 135856. DOI:10.1016/j.scitotenv.2019.135856 |
| [49] |
DIAS D P, MARENCO R A. Tree growth, wood and bark water content of 28 Amazonian tree species in response to variations in rainfall and wood density[J]. iForest, 2016, 9(3): 445-451. DOI:10.3832/ifor1676-008 |
| [50] |
HERTZBERG M, ASPEBORG H, SCHRADER J, et al. A transcriptional roadmap to wood formation[J]. Proc Natl Acad Sci USA, 2001, 98(25): 14732-14737. DOI:10.1073/pnas.261293398 |
| [51] |
ZHU X L, ZHOU C P, JIA C R, et al. Association of SNP loci and candidate genes for growth and wood density in Eucalyptus urophylla× E. tereticornis[J]. J Nanjing For Univ (Nat Sci), 2021, 45(4): 143-150. 朱显亮, 周长品, 贾翠蓉, 等. 尾细桉生长和木材密度关联SNP挖掘与候选基因定位[J]. 南京林业大学学报(自然科学版), 2021, 45(4): 143-150. DOI:10.12302/j.issn.1000-2006.202004003 |
| [52] |
XU J, WANG X Y, GUO W Z. The cytochrome P450 superfamily: Key players in plant development and defense[J]. J Integr Agric, 2015, 14(9): 1673-1686. DOI:10.1016/S2095-3119(14)60980-1 |
| [53] |
CUI H T, JIANG X, ZHANG T J, et al. The research progress of plant cytochrome P450 family[J]. Chin J Grassland, 2020, 42(5): 173-180. 崔会婷, 蒋旭, 张铁军, 等. 植物CYP450家族研究进展[J]. 中国草地学报, 2020, 42(5): 173-180. DOI:10.16742/j.zgcdxb.20190182 |
| [54] |
HE L H, ZHAO S J, HU Z B. Gene and function research progress of plant cytochrome P450s[J]. Pharm Biotechnol, 2008, 15(2): 142-147. 贺丽虹, 赵淑娟, 胡之璧. 植物细胞色素P450基因与功能研究进展[J]. 药物生物技术, 2008, 15(2): 142-147. DOI:10.19526/j.cnki.1005-8915.2008.02.015 |
| [55] |
CANKAR K, VAN HOUWELINGEN A, GOEDBLOED M, et al. Valencene oxidase CYP706M1 from Alaska cedar (Callitropsis nootkatensis)[J]. FEBS Lett, 2014, 588(6): 1001-1007. DOI:10.1016/j.febslet.2014.01.061 |
| [56] |
RALSTON L, YU O. Metabolons involving plant cytochrome P450s[J]. Phytochem Rev, 2006, 5(2/3): 459-472. DOI:10.1007/s11101-006-9014-4 |
| [57] |
ZHAO Y D, HULL A K, GUPTA N R, et al. Trp-dependent auxin biosynthesis in Arabidopsis: Involvement of cytochrome P450s CYP79B2 and CYP79B3[J]. Genes Dev, 2002, 16(23): 3100-3112. DOI:10.1101/gad.1035402 |
| [58] |
ZHANG C B, ZHAO L M, ZHAO H K, et al. Advances in plant protein kinase[J]. Biotechnol Bull, 2011, 27(10): 17-23. 张春宝, 赵丽梅, 赵洪锟, 等. 植物蛋白激酶研究进展[J]. 生物技术通报, 2011, 27(10): 17-23. DOI:10.13560/j.cnki.biotech.bull.1985.2011.10.019 |
| [59] |
KUSHWAH S, BANASIAK A, NISHIKUBO N, et al. Arabidopsis XTH4 and XTH9 contribute to wood cell expansion and secondary wall formation[J]. Plant Physiol, 2020, 182(4): 1946-1965. DOI:10.1104/pp.19.01529 |
| [60] |
DU J, GERTTULA S, LI Z H, et al. Brassinosteroid regulation of wood formation in poplar[J]. New Phytol, 2020, 225(4): 1516-1530. DOI:10.1111/nph.15936 |
| [61] |
LEE J, HAN S, LEE H Y, et al. Brassinosteroids facilitate xylem differentiation and wood formation in tomato[J]. Planta, 2019, 249(5): 1391-1403. DOI:10.1007/s00425-019-03094-6 |
| [62] |
NAGATA N, ASAMI T, YOSHIDA S. Brassinazole, an inhibitor of brassinosteroid biosynthesis, inhibits development of secondary xylem in cress plants (Lepidium sativum)[J]. Plant Cell Physiol, 2001, 42(9): 1006-1011. DOI:10.1093/pcp/pce122 |
 2024, Vol. 32
2024, Vol. 32


