2. 中国科学院大学, 北京 100049;
3. 黑龙江省农业科学院, 哈尔滨 150086
2. University of Chinese Academy of Sciences, Beijing 100049, China;
3. Heilongjiang Academy of Agricultural Sciences, Harbin 150086, China
栽培大豆(Glycine max)起源于中国黄淮海地区,由野生大豆(G. soja)驯化而来[1]。与水稻(Oryza sativa)、小麦(Triticum aestivum)等粮食作物一样,大豆以种子为主要食用对象。因其含有丰富的蛋白质(约40%)和油脂(约20%)[2–3],大豆已成为人类食品和动物饲料中不可替代的植物蛋白和重要食用油来源。目前,全球近50%的植物蛋白质和植物油脂来源于大豆(www.soystats.com)。然而,面对全球快速增长的人口问题及养殖业的快速发展,选育高产优质的大豆品种已成为育种家们的首要目标[4–5]。
大豆种子蛋白和油脂含量是多基因控制的数量性状,同时还受环境因素的影响。多因素和多种调控途径共同决定了大豆种子的蛋白和油脂含量,虽然目前有一些研究报道了调控大豆蛋白油脂含量的基因位点,但关键基因的功能鉴定和调控网络还不清晰。此外,蛋白质和油脂含量间呈现此消彼长的关系[2],这也成为限制大豆蛋白和油脂含量共同提高的主要因素。解析大豆种子蛋白和油脂含量形成的分子机理,揭示两者之间的协同调控网络,是创制高产优质大豆品种的重要理论基础[6]。近年来,随着大豆基因组学和现代分子生物学研究的不断深入,利用基因定向改良等手段,精确调控大豆种子蛋白和油脂含量,以实现高产优质大豆品种的育种目标。本文针对大豆蛋白和油脂品质性状的分子调控机制进行综述,并讨论了目前大豆蛋白和油脂性状分子研究面临的挑战和前景,以期为大豆种子品质定向改良提供理论参考。
1 大豆种子贮藏物质的组成种子发育是大豆生长发育的关键一环,决定了产量的高低和品质的优劣[7]。在大豆的生长发育过程中,叶片等“源”器官经过光合作用产生蔗糖并运输到种子中。种子发育初期首先是淀粉积累,到种子发育中后期,淀粉水解以提供大量能量和原料,合成蛋白质和油脂。成熟种子中淀粉含量占籽粒干重的1%~3%,此外,还含有26%~30%的碳水化合物。碳水化合物组成复杂,其中,包括棉子糖、水苏糖在内的大豆低聚糖占比最高。蛋白质和油脂是大豆种子的主要组成部分,分别约占大豆组成的40%和20%[8]。蛋白质和油脂含量也是大豆用途和品质的主要决定因素。然而,大豆种子中蛋白质含量和油脂含量呈负相关关系,二者之间存在底物竞争关系[2]。
2 大豆种子油脂含量的调控 2.1 油脂生物合成关键酶大豆种子的油脂累积与多数植物种子相似,受到脂肪酸(FAs)生物合成、三酰基甘油(TAGs)组装以及油脂和其他细胞代谢物之间碳分配过程这3方面的影响[9–10]。脂肪酸(FAs)在质体中合成,此过程受到乙酰辅酶A羧化酶(ACCase)和3-酮脂酰-酰基载体蛋白合酶(KAS)等多种关键酶的调控。积累的FAs随后转移至内质网,经过G3P酰基转移酶(GPAT)、溶血磷脂酸酰基转移酶(LPAAT)、磷酸酶(PAP)和二酰基甘油酰基转移酶(DGAT)的酰化和酯化(肯尼迪途径),完成TAGs的组装[11]。其中,大豆中TAGs产生的主要途径是由磷脂酰胆碱(PC)转化为1,2-sn-二酰基甘油(DAG),DAG再酰化产生TAGs[12]。在碳分配方面,丙酮酸脱氢酶激酶(PDHK)和胞质d-葡萄糖-6-磷酸脱氢酶(Glu6PDH)影响FAs的合成进而影响种子油脂含量[13–14](图1)。

|
图 1 大豆油脂合成的转录调控模块 Fig. 1 Transcriptional regulatory modules of soybean oil synthesis |
在种子胚胎发育后期,脂肪酶介导TAG转变导致大豆种子脂肪酸含量下降,寡糖、棉子糖和水苏糖含量升高。研究表明,抑制脂肪酶介导TAG的转变可以增加脂肪酸含量,减少不可消化的低聚糖的存在。大豆中的GmSDP1基因与拟南芥(Arabidopsis thaliana)脂肪酶基因同源,编码TAG脂肪酶。GmSDP1介导TAG转换的同时影响脂肪酸的组成,抑制SDP1基因的表达不仅能够促进种子油分的积累,增加油酸含量,降低亚油酸的含量还能使蛋白质含量呈上升趋势[15–16]。
除了增加油脂的合成与累积,改变大豆中脂肪酸的组成比例,提升油脂利用率也是目前的育种策略之一。大豆的油脂以酰基甘油(TAGs)形式储存在种子中[11],具体可分为棕榈酸(16∶0)、硬脂酸(18∶0)、油酸(18∶1)、亚油酸(18∶2)和亚麻酸(18∶3)这5种脂肪酸[3], 其中,亚油酸和亚麻酸约占68%,属于多不饱和脂肪酸(PUFAs),易于氧化,过多的PUFAs会降低大豆油的氧化稳定性,影响大豆品质[3]。
2.2 转录因子转录因子在种子油脂合成与累积过程中也发挥着重要作用。目前,大豆种子中已鉴定出多个油脂合成相关的重要转录因子,如GmDofs、GmMYB73、LECs (Leaf cotyledon)和WRI1 (Wrinkled1)等(图1)[17–22]。
GmDof4 (DNA binding with one finger 4)和GmDof11是2个油脂合成相关的转录因子,参与FAs的合成调控。GmDof4和GmDof11分别激活了ACCase和长链脂酰辅酶A合成酶(LACS)基因的表达,提高转基因拟南芥种子中总脂肪酸和脂质的含量。Zhang等[23]将GmDOf4基因转入椭圆小球藻, GmDof4能够显著促进其油分累积。此外,GmDof4和GmDof11能够直接结合储存蛋白基因CRA1的启动子,下调储存蛋白基因的表达[17]。
在大豆中特异表达的GmMYB73属于MYB家族的转录因子,参与TAGs的合成调控。研究表明,GmMYB73能够与bHLH型转录因子GL3 (Glabra 3)和EGL3 (Enhancer of glabra 3)互作,抑制油分累积的负调控因子GL2 (Glabra 2)的表达,进而解除GL2对磷脂酶PLDα1的抑制。解除抑制的PLDα1促进PC水解并最终合成DAG和TAG[18]。此外, GqOil2上的候选基因GmOLEO1编码油球蛋白能够影响TAG的代谢,促进油分的积累[24]。
GmLEC2a是含有B3结构域的转录因子。GmLEC2a基因位于第20号染色体上,与拟南芥LEC2同源。拟南芥的GmLEC2a过表达株系的TAG含量增加34%,长链脂肪酸组成增加4%。Manan等[19]将GmLEC2a在大豆毛状根中异位表达,结果多个与油脂合成相关的转录因子表达上调,包括GmLEC1、GmFUS3、GmABI3 (ABA insentive3)、GmDof11和GmWRI1。
WRI1是编码AP2/乙烯响应元件结合蛋白(APE TALA2/EREBP)家族的一个转录因子。在GmWRI1的3个同源基因GmWRI1a、GmWRI1b和GmWRI1c中,GmWRI1a主要在大豆种子中表达。GmWRI1a过表达大豆转基因株系种子的总油量和脂肪酸含量增加。此外,GmWRI1a能够与保守的AW-box顺式作用元件特异性相互作用,控制大豆油分相关生物合成基因的表达[25]。
转录因子GmbZIP123可以直接结合蔗糖转运体基因SUC1、SUC5和细胞壁转化酶基因cwINV1、cwINV3和cwINV6的启动子,并促进这些基因的表达,控制糖从光自养组织进入种子,参与大豆种子中脂质积累的调控[26]。此外,Hu等[27]鉴定了1个种子驯化过程中的选择基因GmZF351,其编码串联CCCH锌指蛋白。GmZF351能够激活脂质生物合成相关基因GmBCCP2 (biotin carboxyl carrier protein 2)、GmKASIII、GmDGAT1 (diacylglycerol-O-acyl-transferase 1)和GmOLEO2 (oleosin 2)的表达,增强WRI1活性,正向调节脂质的生物合成[28]。锌指蛋白GmZF392同样能够提高大豆种子的油脂含量, 一方面GmZF392能够与启动子区富含TG和TA的双元元件结合,激活脂质生物合成途径中基因的表达;另一方面,GmZF392与GmZF351互作,协同促进下游基因的表达。不仅如此,NF-Y类的核因子GmNFYA分别通过直接和间接调控促进GmZF392和GmZF351基因的表达,提高转基因大豆种子中的含油量[29](表1)。
| 表 1 种子油脂含量的主要调控因子 Table 1 Major regulatory factors that control seed oil content |
大豆种子蛋白质根据功能可分为11组,其中,含量最丰富的是种子储藏蛋白。储藏蛋白主要由球蛋白(11S)和β-半球蛋白(7S)组成[30–31],两者共同影响着大豆蛋白的含量与品质[32–34]。大豆种子球蛋白和β-半球蛋白分别为六亚基和三亚基聚合蛋白,分别由多基因Gy1-Gy7和CG1-CG15编码[35–36],其中,7S上的β亚基编码基因CG4由1个位于第20号染色体上的显性位点qBSC-1控制,影响大豆蛋白的质量和加工性能[37]。相较于大豆种子油脂,对控制大豆种子蛋白的基因功能鉴定较少。Han等[38]利用整合转录组学和蛋白质组学特征对1个野生大豆品系的染色体片段替换系(CSSL)和亲本系进行分析, 鉴定出27个差异表达基因(DEGs)和23个差异积累蛋白(DAPs)调控种子储藏蛋白。这些基因编码的蛋白参与光合作用、蛋白质加工、蛋白质分选和储存蛋白质积累过程(图2)。
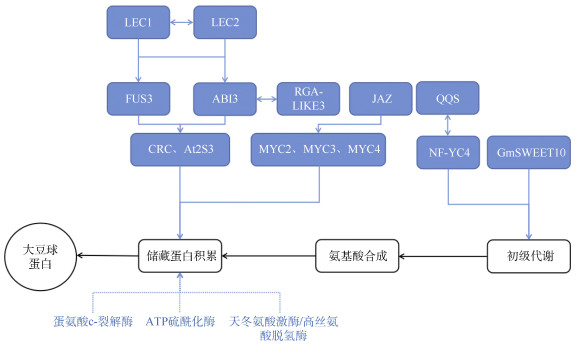
|
图 2 大豆蛋白合成的转录调控模块 Fig. 2 Transcriptional regulatory module of soybean protein synthesis |
种子蛋白质和油脂合成所需的原料主要来源于糖类物质的分解。糖的分配影响胚胎的发育也调节脂肪酸的生物合成和蛋白质的生物合成[39–40]。位于15号染色体上的糖转运体基因Glyma.15G049200 (GmSWEET10)调控着种子蛋白质和油脂的积累,该基因编码的蛋白定位于质膜,属于SWEET家族成员[41]。此外,Glyma.15G049200基因存在等位基因GmSWEET10a和GmSWEET10b,两者功能冗余, 主要表达于种皮的薄壁组织和表皮中[42],可能通过影响糖从母体种皮到子代胚的传递来调节油脂和蛋白质的积累[43]。
碳源和氮源的分配影响着氨基酸和脂肪酸的合成进而影响种子蛋白质和油脂的含量。Li等[44]报道在拟南芥中特有的“孤儿基因”QQS能够与转录因子NF-YC4相互作用调控碳和氮分配促进种子的蛋白质合成。此外,NF-YC4在植物中广泛存在,其同源物能够与QQS蛋白结合互作,QQS能够调控拟南芥、大豆等物种的碳和氮分配,从而影响种子蛋白质的合成。大豆储存蛋白最初在内质网上合成,然后遵循高尔基体介导的运输途径沉积至蛋白质储存液泡(PSV)。Wei等[45]报道在大豆子叶发育过程中,小的GTPase编码基因GmRab5a及其鸟嘌呤核苷酸交换因子(GEFs),参与调控大豆储存蛋白的运输,影响蛋白质的积累。
一些酶也调控着大豆蛋白质的组成和积累。研究表明,蛋氨酸c-裂解酶(MGL)调控大豆种子中甲基蛋氨酸的积累[46]。蛋氨酸和半胱氨酸分别为人体必须氨基酸和条件必需氨基酸,过表达的ATP硫酰化酶(ATPS)能够促进大豆中这2种氨基酸的合成, 从而提升大豆蛋白品质[47]。磷酸烯醇丙酮酸羧激酶(PEPC)活性在高蛋白品种中高于低蛋白品种,可作为大豆育种中的一种生物标志物[48]。Zhang等[49]在qOC-8-1 (qWSPC-8-1)株系中鉴定出1个对油脂和水溶性蛋白的合成起反作用的Glyma.08G107800基因,该基因的表达与大豆的油类合成呈负相关,与水溶性蛋白的合成呈正相关,Glyma.08G107800编码一种双功能酶:天冬氨酸激酶/高丝氨酸脱氢酶(AK-HSDH),该酶对天冬氨酸家族(赖氨酸、苏氨酸、甲硫氨酸和异亮氨酸)的氨基酸合成非常重要。
种子蛋白质的合成与积累除了由其合成代谢和组分构成等因素决定,还受到多种转录因子的共同调控。目前,对于控制大豆种子蛋白合成的转录因子鉴定较少,已报道的相关转录因子多集中于拟南芥中。拟南芥中鉴定的FUS3、LEC2和ABI3 (ABA insentive3)是植物特异的B3类转录因子,能够以直接调控或间接调控的方式影响储藏蛋白基因At2S3的表达,其中,FUS3和LEC2存在部分功能冗余[50–51]。核因子LEC1是植物种子发育的核心调控因子,参与种子发育的多个生物学进程。Kagaya等[52]研究表明,LEC1通过控制FUS3和ABI3协同调控种子储藏蛋白基因CRC和At2S3的表达,从而影响种子储藏蛋白的积累。Chern等[53]在菜豆(Phaseolus vulgaris)中鉴定出bZIP类转录因子ROM1是一种DNA结合位点依赖的抑制因子。ROM1与转录激活因子PvALF拮抗,抑制发育过程中种子蛋白合成基因的转录。此外,转录因子与激素之间相互作用同样影响着种子储藏蛋白的积累。赤霉素信号负调控因子RGA-LIKE3作为ABI3的共激活因子,在种子成熟阶段促进储藏蛋白的生物合成[54];茉莉酸信号转导中的抑制蛋白JAZ能够直接靶标bHLH类转录因子MYC2、MYC3和MYC4,协同调控种子储藏蛋白的积累[55]。近期研究表明,在第20号染色体上存在1个与大豆种子蛋白含量呈负相关的基因Glyma.20G85100,其编码的蛋白中CCT结构域存在1个转座子的插入,从而影响该基因的表达,同时用RNAi干扰抑制Glyma.20G85100基因,能够显著提升种子蛋白含量,具体机制仍有待研究[56](表2)。
| 表 2 种子蛋白含量的主要调控因子 Table 2 Major regulatory factors that control seed proteins |
大豆种子的油脂、蛋白含量是由多基因控制且易受环境影响的数量性状。Lee等[57]利用限制性片段长度多态性(RFLP)标记对(“Young”×PI416937)和(PI97100דCoker237”)进行分析,在第15 (E连锁群)、18 (G连锁群)、12 (H连锁群)和9 (K连锁群)号染色体上存在多种蛋白和油脂相关的共同标记。Liang等[58]利用群体(Jindou23×Huibuzhi)分别检测出6个蛋白和11个油脂数量性状座位(quantitative trait locus, QTL)。从2个群体(Magellan×PI 438489B)和(Magellan×PI 567516C)分别获得216和156个重组自交系。Pathan等[59]在大豆中分别确定了7个蛋白和6个油脂QTL,其中,第5和6号染色体上存在蛋白质和油脂共同的QTL。Li等[60]则对长江、淮河流域的大豆种子进行多位点全基因组关联分析, 鉴定出10个新的与油脂或蛋白相关的QTLs,还分别鉴定出55个与种子蛋白和51个与油脂相关的候选基因。Zhang等[49]对211份大豆种质进行种子组成的全基因组关联研究(genome-wide association studies, GWAS)分析,分别鉴定出蛋白质(3个)、脂肪(4个)和水溶性蛋白质(5个)相关的QTL,其中, qOC-8-1 (qWSPC-8-1)对油脂和水溶性蛋白的合成起反比作用。GqOil20是1个环境稳定的QTL,与油脂含量显著相关,占多种环境下种子油脂总表型变异量的23.70%[24]。
通过对不同群体进行QTL和GWAS分析,已报道的与大豆蛋白和油脂相关的QTLs分别有248和327个,分布于20条染色体上(http://www.soybase.org, 2022-07) (表3)。大豆种子蛋白相关的QTLs位于第6、9、18和20号染色体上较多,其中,第20号染色体上的蛋白相关的QTL与GWAS QTL数目均最多,最高可解释65%的表型变异[61]。大豆种子油脂相关的QTLs位于第5、15、18、19和20号染色体上较多,其中,第15、18和19号染色体上的QTL数目均为 23个。
5 讨论和展望大豆是世界粮食安全的重要组成部分,大豆蛋白优质[62],能够提供人体所不能合成的必需氨基酸,同时大豆油脂中饱和脂肪酸较少,胆固醇含量低,有益于人体健康[63–64]。长期以来,大豆已成为人类食品和饲料中不可替代的植物蛋白和食用油来源。面对世界人口激增、耕地面积短缺和多变的自然气候,培育出能够适应未来环境变化的优质大豆品种已成为迫切需要。提高大豆油脂和蛋白质含量更是一直以来的育种目标。
为培育高产优质的大豆品种,通常需要将多个优良品质性状聚合,但大豆油脂和蛋白质含量受多个数量位点的控制,且调控途径相互串扰,传统育种技术很难克服这个限制。有研究认为大豆蛋白与油脂之间存在一定的底物竞争,两者合成所需的原料均来源于糖类物质的分解[65–66]。打破蛋白与油脂间的负相关性以及维持多性状的平衡关系是实现大豆品质改良的关键。目前,油脂蛋白合成的调控研究主要为合成途径中关键酶[67–69]及一些转录因子[70–71], 仍有大量的种子蛋白油脂含量的位点需要克隆和功能验证,从而进一步完善大豆蛋白和油脂基因的合成代谢网络。
现代栽培大豆由野生大豆驯化而来,长期的驯化历程使植物朝着更有益于人类社会生产的方向发展,但这也导致植物大量优异农艺性状丢失,遗传多样性降低。如何在避免其他优异农艺性状丢失的同时选择目标性状,有研究者认为从头驯化可能是另一种途径[72–74]。野生大豆具有高蛋白低油脂的性状,经过千年的驯化,如今的栽培大豆油脂含量高,蛋白含量低,抗逆性差。通过对野生大豆从头驯化或再驯化,针对性的选择高油脂和蛋白含量等位基因,清除有害变异,减少或避免其他优异基因的丢失,也可能会实现大豆高产优质的育种目标[75]。
现代分子生物学、基因组学的快速发展,促进了蛋白和油脂含量调控基因的挖掘和调控网络的解析。同时,随着基因定向编辑[76–77]、大豆快速遗传转化[78–79]等技术的进一步突破,也会加速大豆品质性状的精准改良,最终实现大豆产量和品质的共同提高。
| [1] |
LI Y H, GUAN R X, LIU Z X, et al. Genetic structure and diversity of cultivated soybean (Glycine max (L. ) Merr. ) landraces in China[J]. Theor Appl Genet, 2008, 117(6): 857-871. DOI:10.1007/s00122-008-0825-0 |
| [2] |
CHAUDHARY J, PATIL G B, SONAH H, et al. Expanding omics resources for improvement of soybean seed composition traits[J]. Front Plant Sci, 2015, 6: 1021. DOI:10.3389/fpls.2015.01021 |
| [3] |
CLEMENTE T E, CAHOON E B. Soybean oil: Genetic approaches for modification of functionality and total content[J]. Plant Physiol, 2009, 151(3): 1030-1040. DOI:10.1104/pp.109.146282 |
| [4] |
GODFRAY H C J, BEDDINGTON J R, CRUTE I R, et al. Food security: The challenge of feeding 9 billion people[J]. Science, 2010, 327(5967): 812-818. DOI:10.1126/science.1185383 |
| [5] |
TILMAN D, BALZER C, HILL J, et al. Global food demand and the sustainable intensification of agriculture[J]. Proc Natl Acad Sci USA, 2011, 108(50): 20260-20264. DOI:10.1073/pnas.1116437108 |
| [6] |
TIAN Z X, WANG J W, LI J Y, et al. Designing future crops: Challenges and strategies for sustainable agriculture[J]. Plant J, 2021, 105(5): 1165-1178. DOI:10.1111/tpj.15107 |
| [7] |
DU J, WANG S D, HE C M, et al. Identification of regulatory networks and hub genes controlling soybean seed set and size using RNA sequencing analysis[J]. J Exp Bot, 2017, 68(8): 1955-1972. DOI:10.1093/jxb/erw460 |
| [8] |
ZHANG H Y, HU Z B, YANG Y M, et al. Transcriptome profiling reveals the spatial-temporal dynamics of gene expression essential for soybean seed development[J]. BMC Genom, 2021, 22(1): 453. DOI:10.1186/s12864-021-07783-z |
| [9] |
KELLY A A, VAN ERP H, QUETTIER A L, et al. The sugardependent1 lipase limits triacylglycerol accumulation in vegetative tissues of Arabidopsis[J]. Plant Physiol, 2013, 162(3): 1282-1289. DOI:10.1104/pp.113.219840 |
| [10] |
ZHAO C G, LIU D, LI F L, et al. Advances in research on seed oil biosynthesis and basal metabolism[J]. Seeds, 2010, 29(4): 56-62. 赵翠格, 刘頔, 李凤兰, 等. 植物种子油脂的生物合成及代谢基础研究进展[J]. 种子, 2010, 29(4): 56-62. DOI:10.16590/j.cnki.1001-4705.2010.04.043 |
| [11] |
CHAPMAN K D, OHLROGGE J B. Compartmentation of triacylglycerol accumulation in plants[J]. J Biol Chem, 2012, 287(4): 2288-2294. DOI:10.1074/jbc.R111.290072 |
| [12] |
BATES P D, DURRETT T P, OHLROGGE J B, et al. Analysis of acyl fluxes through multiple pathways of triacylglycerol synthesis in developing soybean embryos[J]. Plant Physiol, 2009, 150(1): 55-72. DOI:10.1104/pp.109.137737 |
| [13] |
WAKAO S, ANDRE C, BENNING C. Functional analyses of cytosolic glucose-6-phosphate dehydrogenases and their contribution to seed oil accumulation in Arabidopsis[J]. Plant Physiol, 2008, 146(1): 277-288. DOI:10.1104/pp.107.108423 |
| [14] |
MARILLIA E F, MICALLEF B J, MICALLEF M, et al. Biochemical and physiological studies of Arabidopsis thaliana transgenic lines with repressed expression of the mitochondrial pyruvate dehydrogenase kinase[J]. J Exp Bot, 2003, 54(381): 259-270. DOI:10.1093/jxb/erg020 |
| [15] |
AZNAR-MORENO J A, MUKHERJEE T, MORLEY S A, et al. Suppression of SDP1 improves soybean seed composition by increasing oil and reducing undigestible oligosaccharides[J]. Front Plant Sci, 2022, 13: 863254. DOI:10.3389/fpls.2022.863254 |
| [16] |
KANAI M, YAMADA T, HAYASHI M, et al. Soybean (Glycine max L. ) triacylglycerol lipase GmSDP1 regulates the quality and quantity of seed oil[J]. Sci Rep, 2019, 9(1): 8924. DOI:10.1038/s41598-019-45331-8 |
| [17] |
WANG H W, ZHANG B, HAO Y J, et al. The soybean Dof-type transcription factor genes, GmDof4 and GmDof11, enhance lipid content in the seeds of transgenic Arabidopsis plants[J]. Plant J, 2007, 52(4): 716-729. DOI:10.1111/j.1365-313x.2007.03268.x |
| [18] |
LIU Y F, LI Q T, LU X, et al. Soybean GmMYB73 promotes lipid accumulation in transgenic plants[J]. BMC Plant Biol, 2014, 14(1): 73. DOI:10.1186/1471-2229-14-73 |
| [19] |
MANAN S, AHMAD M Z, ZHANG G Y, et al. Soybean LEC2 regulates subsets of genes involved in controlling the biosynthesis and catabolism of seed storage substances and seed development[J]. Front Plant Sci, 2017, 8: 1604. DOI:10.3389/fpls.2017.01604 |
| [20] |
BAUD S, MENDOZA M S, TO A, et al. WRINKLED1 specifies the regulatory action of LEAFY COTYLEDON2 towards fatty acid metabolism during seed maturation in Arabidopsis[J]. Plant J, 2007, 50(5): 825-838. DOI:10.1111/j.1365-313x.2007.03092.x |
| [21] |
CERNAC A, BENNING C. WRINKLED1 encodes an AP2/EREB domain protein involved in the control of storage compound biosynthesis in Arabidopsis[J]. Plant J, 2004, 40(4): 575-585. DOI:10.1111/j.1365-313X.2004.02235.x |
| [22] |
MU J Y, TAN H L, ZHENG Q, et al. LEAFY COTYLEDON1 is a key regulator of fatty acid biosynthesis in Arabidopsis[J]. Plant Physiol, 2008, 148(2): 1042-1054. DOI:10.1104/pp.108.126342 |
| [23] |
ZHANG J H, HAO Q, BAI L L, et al. Overexpression of the soybean transcription factor GmDof4 significantly enhances the lipid content of Chlorella ellipsoidea[J]. Biotechnol Biofuels, 2014, 7(1): 128. DOI:10.1186/s13068-014-0128-4 |
| [24] |
ZHANG D, ZHANG H Y, HU Z B, et al. Artificial selection on GmOLEO1 contributes to the increase in seed oil during soybean domestication[J]. PLOS Genet, 2019, 15(7): e1008267. DOI:10.1371/journal.pgen.1008267 |
| [25] |
CHEN L, ZHENG Y H, DONG Z M, et al. Soybean (Glycine max) WRINKLED1 transcription factor, GmWRI1a, positively regulates seed oil accumulation[J]. Mol Genet Genom, 2018, 293(2): 401-415. DOI:10.1007/s00438-017-1393-2 |
| [26] |
SONG Q X, LI Q T, LIU Y F, et al. Soybean GmbZIP123 gene enhances lipid content in the seeds of transgenic Arabidopsis plants[J]. J Exp Bot, 2013, 64(14): 4329-4341. DOI:10.1093/jxb/ert238 |
| [27] |
HU X, ZUO J F. The CCCH zinc finger family of soybean (Glycine max L. ): Genome-wide identification, expression, domestication, GWAS and haplotype analysis[J]. BMC Genom, 2021, 22(1): 511. DOI:10.1186/s12864-021-07787-9 |
| [28] |
LI Q T, LU X, SONG Q X, et al. Selection for a Zinc-finger protein contributes to seed oil increase during soybean domestication[J]. Plant Physiol, 2017, 173(4): 2208-2224. DOI:10.1104/pp.16.01610 |
| [29] |
LU L, WEI W, LI Q T, et al. A transcriptional regulatory module controls lipid accumulation in soybean[J]. New Phytol, 2021, 231(2): 661-678. DOI:10.1111/nph.17401 |
| [30] |
GUAN R X, CHANG R Z, QIU L J, et al. Analysis of protein subunit 7S/11S constitution and allergen lacking of soybean [Glycine max (L. ) Merrill] cultivars[J]. Acta Agron Sin, 2004, 30(11): 1076-1079. 关荣霞, 常汝镇, 邱丽娟, 等. 栽培大豆蛋白亚基11S/7S组成及过敏蛋白缺失分析[J]. 作物学报, 2004, 30(11): 1076-1079. DOI:10.3321/j.issn:0496-3490.2004.11.002 |
| [31] |
AO Y, WANG A, WU Q, et al. Research progress on metabolic pathways of seed protein and related regulation mechanism in soybean[J]. Soybean Sci, 2018, 37(5): 794-802. 敖雁, 王安, 吴启, 等. 大豆籽粒蛋白代谢途径及相关调控机制研究进展[J]. 大豆科学, 2018, 37(5): 794-802. DOI:10.11861/j.issn.1000-9841.2018.05.0794 |
| [32] |
KRISHNAN H B, OEHRLE N W, NATARAJAN S S. A rapid and simple procedure for the depletion of abundant storage proteins from legume seeds to advance proteome analysis: A case study using Glycine max[J]. Proteomics, 2009, 9(11): 3174-3188. DOI:10.1002/pmic.200800875 |
| [33] |
RUÍZ-HENESTROSA V P, SÁNCHEZ C C, ESCOBAR M D M Y, et al. Interfacial and foaming characteristics of soy globulins as a function of pH and ionic strength[J]. Colloids Surfaces Physicochem Engin Asp, 2007, 309(1/2/3): 202-215. DOI:10.1016/j.colsurfa.2007.01.030 |
| [34] |
ZHOU R B, ZHOU B. The structure and functional properties of soybean 7S and 11S globulin proteins[J]. J Chin Cereals Oils Assoc, 1998, 13(6): 439-442. 周瑞宝, 周兵. 大豆7S和11S球蛋白的结构和功能性质[J]. 中国粮油学报, 1998, 13(6): 439-442. DOI:10.3321/j.issn:1003-0174.1998.06.011 |
| [35] |
BEILINSON V, CHEN Z, SHOEMAKER R, et al. Genomic organization of glycinin genes in soybean[J]. Theor Appl Genet, 2002, 104(6): 1132-1140. DOI:10.1007/s00122-002-0884-6 |
| [36] |
TSUBOKURA Y, HAJIKA M, KANAMORI H, et al. The β-conglycinin deficiency in wild soybean is associated with the tail-to-tail inverted repeat of the α-subunit genes[J]. Plant Mol Biol, 2012, 78(3): 301-309. DOI:10.1007/s11103-011-9865-y |
| [37] |
WANG J, LIU L, GUO Y, et al. A dominant locus, qBSC-1, controls β subunit content of seed storage protein in soybean (Glycine max (L. ) Merri. )[J]. J Integr Agric, 2014, 13(9): 1854-1864. DOI:10.1016/s2095-3119(13)60579-1 |
| [38] |
HAN X, LI J P, ZHAO Y B, et al. Integrated transcriptomic and proteomic characterization of a chromosome segment substitution line reveals a new regulatory network controlling the seed storage profile of soybean[J]. Food Energy Secur, 2022, 11(2): e381. DOI:10.1002/fes3.381 |
| [39] |
WEBER H, BORISJUK L, WOBUS U. Molecular physiology of legume seed development[J]. Annu Rev Plant Biol, 2005, 56(1): 253-279. DOI:10.1146/annurev.arplant.56.032604.144201 |
| [40] |
HYMOWITZ T, COLLINS F I, PANCZNER J, et al. Relationship between the content of oil, protein, and sugar in soybean seed[J]. Agron J, 1972, 64(5): 613-616. DOI:10.2134/agronj1972.00021962006400050019x |
| [41] |
MIAO L, YANG S N, ZHANG K, et al. Natural variation and selection in GmSWEET39 affect soybean seed oil content[J]. New Phytol, 2020, 225(4): 1651-1666. DOI:10.1111/nph.16250 |
| [42] |
WANG S D, LIU S L, WANG J, et al. Simultaneous changes in seed size, oil content and protein content driven by selection of SWEET homologues during soybean domestication[J]. Nation Sci Rev, 2020, 7(11): 1776-1786. DOI:10.1093/nsr/nwaa110 |
| [43] |
ZHANG H, GOETTEL W, SONG Q, et al. Selection of GmSWEET39 for oil and protein improvement in soybean[J]. PLOS Genet, 2020, 16(11): e1009114. DOI:10.1371/journal.pgen.1009114 |
| [44] |
LI L, ZHENG W G, ZHU Y B, et al. QQS orphan gene regulates carbon and nitrogen partitioning across species via NF-YC interactions[J]. Proc Natl Acad Sci USA, 2015, 112(47): 14734-14739. DOI:10.1073/pnas.1514670112 |
| [45] |
WEI Z Y, PAN T, ZHAO Y Y, et al. The small GTPase Rab5a and its guanine nucleotide exchange factors are involved in post-golgi trafficking of storage proteins in developing soybean cotyledon[J]. J Exp Bot, 2020, 71(3): 808-822. DOI:10.1093/jxb/erz454 |
| [46] |
TESHIMA T, YAMADA N, YOKOTA Y, et al. Suppressed methionine γ-lyase expression causes hyperaccumulation of S-methylmethionine in soybean seeds[J]. Plant Physiol, 2020, 183(3): 943-956. DOI:10.1104/pp.20.00254 |
| [47] |
KIM W S, SUN-HYUNG J, OEHRLE N W, et al. Overexpression of ATP sulfurylase improves the sulfur amino acid content, enhances the accumulation of Bowman-Birk protease inhibitor and suppresses the accumulation of the β-subunit of β-conglycinin in soybean seeds[J]. Sci Rep, 2020, 10(1): 14989. DOI:10.1038/s41598-020-72134-z |
| [48] |
YAMAMOTO N, MASUMURA T, YANO K, et al. Pattern analysis suggests that phospho enol pyruvate carboxylase in maturing soybean seeds promotes the accumulation of protein[J]. Biosci Biotechnol Biochem, 2019, 83(12): 2238-2243. DOI:10.1080/09168451.2019.1648205 |
| [49] |
ZHANG S S, HAO D R, ZHANG S Y, et al. Genome-wide association mapping for protein, oil and water-soluble protein contents in soybean[J]. Mol Genet Genom, 2021, 296(1): 91-102. DOI:10.1007/s00438-020-01704-7 |
| [50] |
KROJ T, SAVINO G, VALON C, et al. Regulation of storage protein gene expression in Arabidopsis[J]. Development, 2003, 130(24): 6065-6073. DOI:10.1242/dev.00814 |
| [51] |
LÜ X Y. The expression GmLEC1 and GmFUS3 during seed development and itʼs relationship with storage protein synthesis in soybean [D]. Jinzhong: Shanxi Agricultural University, 2016. 吕新云. GmLEC1与GmFUS3在大豆种子发育过程中的表达及其与贮藏蛋白合成的关系 [D]. 晋中: 山西农业大学, 2016. |
| [52] |
KAGAYA Y, TOYOSHIMA R, OKUDA R, et al. LEAFY COTYLEDON1 controls seed storage protein genes through its regulation of FUSCA3 and ABSCISIC ACID INSENSITIVE3[J]. Plant Cell Physiol, 2005, 46(3): 399-406. DOI:10.1093/pcp/pci048 |
| [53] |
CHERN M S, EIBEN H G, BUSTOS M M. The developmentally regulated bZIP factor ROM1 modulates transcription from lectin and storage protein genes in bean embryos[J]. Plant J, 1996, 10(1): 135-148. DOI:10.1046/j.1365-313x.1996.10010135.x |
| [54] |
HU Y L, ZHOU L M, YANG Y H, et al. The gibberellin signaling negative regulator RGA-LIKE3 promotes seed storage protein accumulation[J]. Plant Physiol, 2021, 185(4): 1697-1707. DOI:10.1093/plphys/kiaa114 |
| [55] |
GAO C H, QI S H, LIU K G, et al. MYC2, MYC3, and MYC4 function redundantly in seed storage protein accumulation in Arabidopsis[J]. Plant Physiol Biochem, 2016, 108: 63-70. DOI:10.1016/j.plaphy.2016.07.004 |
| [56] |
FLIEGE C E, WARD R A, VOGEL P, et al. Fine mapping and cloning of the major seed protein quantitative trait loci on soybean chromosome 20[J]. Plant J, 2022, 110(1): 114-128. DOI:10.1111/tpj.15658 |
| [57] |
LEE S H, BAILEY M A, MIAN M A R, et al. RFLP loci associated with soybean seed protein and oil content across populations and locations[J]. Theor Appl Genet, 1996, 93(5/6): 649-657. DOI:10.1007/s00224058 |
| [58] |
LIANG H Z, YU Y L, WANG S F, et al. QTL mapping of isoflavone, oil and protein contents in soybean (Glycine max L. Merr. )[J]. Agric Sci China, 2010, 9(8): 1108-1116. DOI:10.1016/s1671-2927(09)60197-8 |
| [59] |
PATHAN S M, VUONG T, CLARK K, et al. Genetic mapping and confirmation of quantitative trait loci for seed protein and oil contents and seed weight in soybean[J]. Crop Sci, 2013, 53(3): 765-774. DOI:10.2135/cropsci2012.03.0153 |
| [60] |
LI S G, XU H F, YANG J Y, et al. Dissecting the genetic architecture of seed protein and oil content in soybean from the Yangtze and Huaihe River Valleys using multi-locus genome-wide association studies[J]. Int J Mol Sci, 2019, 20(12): 3041. DOI:10.3390/ijms20123041 |
| [61] |
SEBOLT A M, SHOEMAKER R C, DIERS B W. Analysis of a quantitative trait locus allele from wild soybean that increases seed protein concentration in soybean[J]. Crop Sci, 2000, 40(5): 1438-1444. DOI:10.2135/cropsci2000.4051438x |
| [62] |
WU Y, ZHOU T T, QIAN K, et al. Research progress on effect of low soy protein diet on patients with chronic kidney disease[J]. Chin Nurs Res, 2018, 32(19): 3012-3015. 武月, 周婷婷, 钱凯, 等. 低大豆蛋白饮食对慢性肾脏病病人影响的研究进展[J]. 护理研究, 2018, 32(19): 3012-3015. DOI:10.3969/j.issn.1009-6493.2018.19.007 |
| [63] |
WANG X Z, JIANG G L, GREEN M, et al. Identification and validation of quantitative trait loci for seed yield, oil and protein contents in two recombinant inbred line populations of soybean[J]. Mol Genet Genom, 2014, 289(5): 935-949. DOI:10.1007/s00438-014-0865-x |
| [64] |
LEAMY L J, ZHANG H Y, LI C B, et al. A genome-wide association study of seed composition traits in wild soybean (Glycine soja)[J]. BMC Genom, 2017, 18(1): 18. DOI:10.1186/s12864-016-3397-4 |
| [65] |
ALLEN D K, OHLROGGE J B, SHACHAR-HILL Y. The role of light in soybean seed filling metabolism[J]. Plant J, 2009, 58(2): 220-234. DOI:10.1111/j.1365-313X.2008.03771.x |
| [66] |
RUUSKA S A, GIRKE T, BENNING C, et al. Contrapuntal networks of gene expression during Arabidopsis seed filling[J]. Plant Cell, 2002, 14(6): 1191-1206. DOI:10.1105/tpc.000877 |
| [67] |
CHAO M N, HU X G, ZHANG J Y, et al. Cloning and functional analysis of promoter of diacylglycerol acyltransferase gene GmDGAT1A in soybean[J]. Acta Agric Boreali-Sin, 2020, 35(4): 27-34. 晁毛妮, 胡喜贵, 张晋玉, 等. 大豆二酰甘油酰基转移酶基因GmDGAT1A启动子的克隆与功能分析[J]. 华北农学报, 2020, 35(4): 27-34. DOI:10.7668/hbnxb.20190821 |
| [68] |
ZHAO J Z. Phospholipase GmPLD and lipid synthetase GmDGAT and GmLPAT from soybean regulate growth and seed oil in transgenic Arabidopsis plants [D]. Nanjing: Nanjing Agricultural University, 2013. 赵江哲. 大豆磷脂酶基因GmPLD和脂合成酶基因GmDGAT, GmLPAT在调控拟南芥生长和种子油含量中的作用 [D]. 南京: 南京农业大学, 2013. |
| [69] |
LI X D, WU G, WU Y H, et al. The accumulation pattern of fatty acids during the development of soybean seeds[J]. Soybean Sci, 2007, 26(4): 506-510. 李晓丹, 吴刚, 武玉花, 等. 大豆种子发育过程中脂肪酸积累模式研究[J]. 大豆科学, 2007, 26(4): 506-510. DOI:10.3969/j.issn.1000-9841.2007.04.011 |
| [70] |
CHEN B B. Functional characterization of diacylglycerol acyltransferase and WRINKLED1 in soybean [D]. Wuhan: Huazhong Agricultural University, 2019. 陈贝贝. 大豆二酰甘油酰基转移酶(DGAT)和转录因子WRINKLED1 (WRI1)功能研究 [D]. 武汉: 华中农业大学, 2019. |
| [71] |
CHEN J L. Expression and regulation of genes related to lipid and protein synthesis and accumulation in soybean [D]. Guilin: Guangxi Normal University, 2020. 陈锦玲. 大豆油脂和蛋白质合成与累积相关基因的表达及调控研究 [D]. 桂林: 广西师范大学, 2020. |
| [72] |
KHAN M Z, ZAIDI S S E A, AMIN I, et al. A CRISPR way for fastforward crop domestication[J]. Trends Plant Sci, 2019, 24(4): 293-296. DOI:10.1016/j.tplants.2019.01.011 |
| [73] |
ZSÖGÖN A, ČERMÁK T, NAVES E R, et al. De novo domestication of wild tomato using genome editing[J]. Nat Biotechnol, 2018, 36(12): 1211-1216. DOI:10.1038/nbt.4272 |
| [74] |
YU H, LIN T, MENG X B, et al. A route to de novo domestication of wild allotetraploid rice[J]. Cell, 2021, 184(5): 1156-1170.e14. DOI:10.1016/j.cell.2021.01.013 |
| [75] |
FERNIE A R, YAN J B. De novo domestication: An alternative route toward new crops for the future[J]. Mol Plant, 2019, 12(5): 615-631. DOI:10.1016/j.molp.2019.03.016 |
| [76] |
CAI Y P, CHEN L, ZHANG Y, et al. Target base editing in soybean using a modified CRISPR/Cas9 system[J]. Plant Biotechnol J, 2020, 18(10): 1996-1998. DOI:10.1111/pbi.13386 |
| [77] |
LACCHINI E, KIEGLE E, CASTELLANI M, et al. CRISPR-mediated accelerated domestication of African rice landraces[J]. PloS One, 2020, 15(3): e0229782. DOI:10.1371/journal.pone.0229782 |
| [78] |
MOU Z M, ZHAO D K. Gene rational design: The dawn of crop breeding[J]. Trends Plant Sci, 2022, 27(7): 633-636. DOI:10.1016/j.tplants.2022.03.007 |
| [79] |
WANG Z, WEI K, XIONG M, et al. Glucan, water-dikinase 1 (GWD1), an ideal biotechnological target for potential improving yield and quality in rice[J]. Plant Biotechnol J, 2021, 19(12): 2606-2618. DOI:10.1111/pbi.13686 |
 2022, Vol. 30
2022, Vol. 30



