2. 中国科学院微生物研究所, 北京 100101;
3. 中国科学院大学, 北京 100049
2. Institute of Microbiology, Chinese Academy of Sciences, Beijing 100101, China;
3. University of Chinese Academy of Sciences, Beijing 100049, China
The genus Fellhanera, belonging to family Pilocarpaceae, in order Lecanorales, class Lecanoro- mycetes was established in 1986 with its type F. fuscatula (Müll. Arg.) Vězda[1-2], including more than 90 species worldwide[3-4]. Fellhanera is the second largest genus among foliicolous lichens next to Porina Acharius[5] and quite diverse in morphological fea- tures[6]. The distinctive traits of this genus are the comparatively small apothecia with a thin margin; paraplectenchymatous excipulum; usually indistinct, branched, and sparsely to densely anastomosing para- physes; Byssoloma-type asci; ellipsoid to cylindrical, transversely septate to muriform ascospores; and pycnidial conidiomata[6]. Fellhanera are mostly folii- colous, common in pan-tropical ecologies.
China had previously reported nine Fellhanera species, which are distributed in Yunnan, Hainan, Hongkong and Taiwan[7-13]. Based on specimens collected from Hainan Province, Fellhanera masto- thallina (Vain.) Lücking & Sérus. is found new to China.
1 Materials and methods 1.1 Specimens and morphologyThe specimens of new record are deposited in the Fungarium of College of Life Sciences, Liaocheng University (LCUF) and Herbarium Mycologicum Aca- demiae Sinicae-Lichenes (HMAS-L). A dissecting microscope (Olympus SZX16) and a light microscope (Olympus BX53) were used for the morphological and anatomical studies. Measurements were taken from mature vertical sections of fruit bodies mounted in water.
1.2 ChemistryAmyloidity of the ascospores was tested using Lugol's solution. Spot tests with K (10% aqueous solution of potassium hydroxide), C (saturated solution of aqueous sodium hypochlorite), and P (saturated solution of p-phenylenediamine in 95% ethyl alcohol) were performed on the thallus surface. The lichen substances were detected and identified by thin-layer chromatography, using solvent C[14-16].
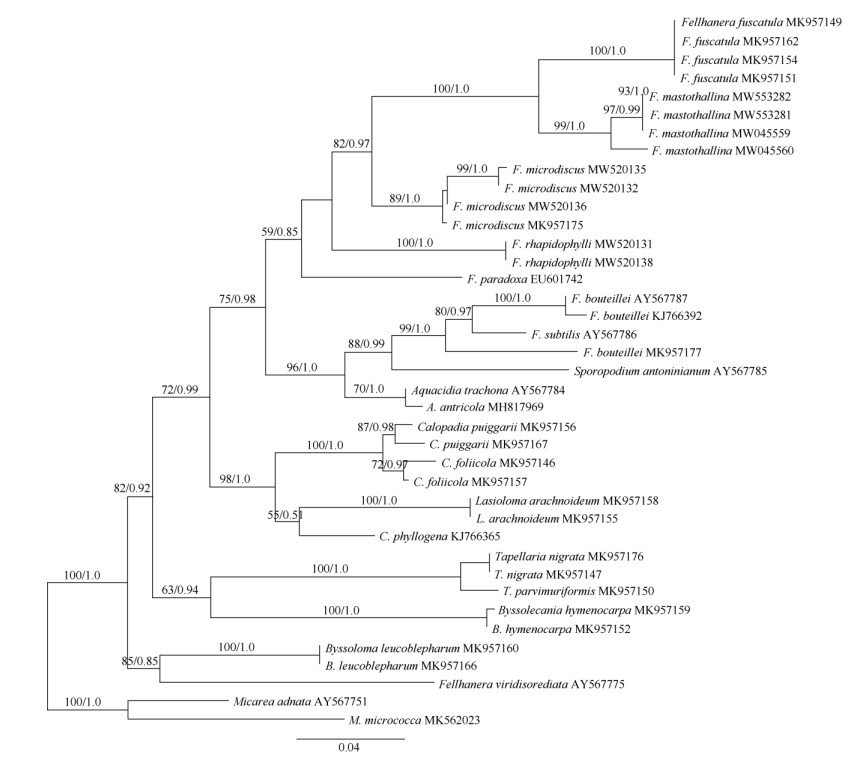
1.3 DNA extraction, amplification, and sequencingGenomic DNA was extracted from ascomata of the specimens using the Hi-DNAsecure Plant Kit (Tiangen, Beijing, China) according to the manufac- turer's protocol. PCR amplification was performed using the mtSSU1 and mtSSU3R primer pair for mtSSU[17]. The 25 μL PCR reaction system containing 1 μL each primer solution (10 μmol/L), 0.5 μL genomic DNA, 10 μL ddH2O, and 12.5 μL 2×Taq PCR Master- Mix (Tiangen, Beijing, China). Thermocycling condi- tions comprised initial denaturation at 95 ℃ (5 min); 35 denaturation cycles at 94 ℃ (45 s), annealing at 50 ℃ (1 min), extension at 72 ℃ (1.5 min) and a final extension at 72 ℃ (10 min). The target product of PCR was affirmed by electrophoresis on 1% agarose gels and sequenced by Biosune Inc. (Shanghai). Nine newly generated sequences were submitted to GenBank. The sequences obtained were evaluated using BLASTn and combined with selected sequences of Pilocarpaceae from GenBank (Fig. 1), Micarea adnata and M. micrococca were used as the outgroup based on Ekman et al. [18]
|
Fig. 1 Maximum likelihood tree of Fellhanera mastothallina and related species within Pilocarpaceae based on the mitochondrial small subunit marker (mtSSU). ML bootstrap values and MCMC posterior probabilities (second value) are displayed above each branch. Branches recovered with ML-BS support ≥70% and BI-PP support ≥0.95 were regarded as strongly supported. GenBank accessions are attached to the sequences. Newly generated sequences are shown in bold. |
Contigs were assembled and edited using the program Geneious v. 6.1.2 (Biomatters Ltd., Auckland, NZ). A total of 39 sequences were aligned using MAFFT v. 7[19]. The program Gblocks v. 0.91b was used to delimit ambiguous regions, implementing all the options for a less stringent selection (http://molevol.cmima.csic.es/castresana/Gblocks_server.html)[20], which yielded final alignment of 670 bp. Maximum likely- hood (ML) and Bayesian inference (BI) were per- formed using the CIPRES Scientific gateway portal (http://www.phylo.org/portal2/)[21]. Maximum likelihood bootstrapping analysis was performed with RAxML- HPC v. 8[22], using the default parameters as imple- mented on the CIPRES, NSF XSEDE resource with bootstrap statistics calculated from 1 000 bootstrap replicates. For the Bayesian analysis, the best substi- tution model was estimated using jModelTest 2.1.6[23]. Based on the results, we used GTR+I+G model. Baye- sian analysis was performed using MrBayes v. 3.2.2 on CIPRES with 2 independent runs, searching for 10 000 000 generations with four independent chains and sampling every 1000th tree[24]. After discarding the burn-in, the remaining 7 500 trees of each run were pooled to calculate a 50% majority rule consensus tree. Generated phylogenetic tree was visualized under Figtree v. 1.4.2[25].
2 Results 2.1 Phylogenetic analysisThe final alignment consisted of 9 newly generated mtSSU sequences and 30 sequences downloaded from NCBI (Fig. 1). The phylogenetic trees obtained from maximum likelihood (ML) and Bayesian inference analysis (BI) exhibited the same topology; we there- fore present only the ML tree. The molecular phylo- geny based on the mitochondrial small subunit marker (mtSSU) of Pilocarpaceae exhibits a well-supported monophyletic lineage containing the genera Bysso- lecania, Byssoloma, Calopadia, Fellhanera, Lasio- loma, Sporopodium and Tapellaria. The tree shows Fellhanera is polyphyletic in its current delimitation. Fellhanera mastothallina is revealed as a sister clade to the type species F. fuscatula. These two species together with F. microdiscus, F. paradoxa and F. rhapidophylli form a monophyletic lineage without good support (BS=59%, PP=0.85). Aquacidia antri- cola, A. trachona, F. bouteillei, F. subtilis and Sporo- podium antoninianum cluster together and get a high support (BS=96%, PP=1.0). While another Fellhanera species, F. viridisorediata shows a close relationship with Byssoloma leucoblepharum.
2.2 TaxonomyFellhanera mastothallina (Vain.) Lücking & Sérus, in Lichenologist 33(3): 192 (2001) Fig. 2
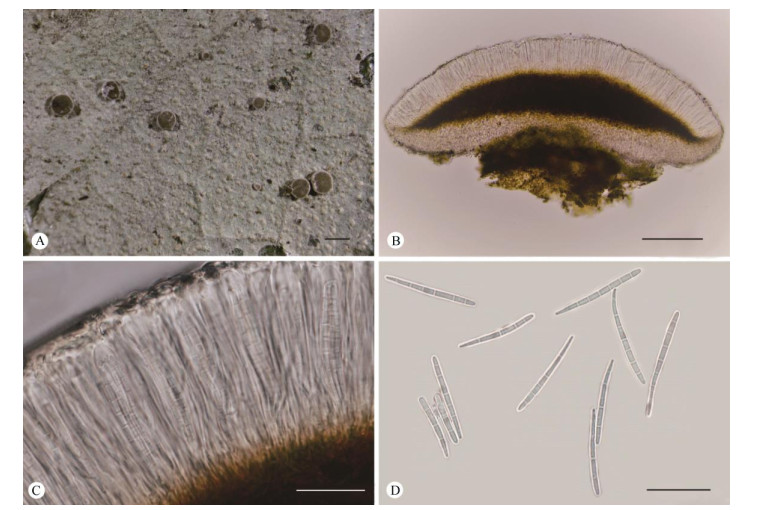
|
Fig. 2 Fellhanera mastothallina (Y.H. Ju HN19458). A: Thallus with apothecia; B: Vertical section of an apothecium; C: Part of a vertical section of an apothecium; D: Ascospores. Bars: A=0.5 mm, B=100 μm, C-D=20 μm |
≡ Bacidia mastothallina Vain., in Ann. Acad. Sci. Fenn., ser. A, 15: 64 (1921)
≡ Bacidina mastothallina (Vain.) Vězda, in Vězda et al., Ann. Naturh. Mus. Wien 99B: 738 (1997)
Type: Philippines, Robinson & Ramos 11900 (TUR-holotype).
Description: Thallus foliicolous, crustose, conti- nuous, verrucose, 10-40 mm across and 10-15 μm thick, greyish-green, rough, often irregular in outline, with light green to orange-red verrucosa of 0.01-0.03 mm diam. Apothecia when mature sessile, rounded, 0.2-0.5 mm diam. and 150-200 μm high; disc plane to slightly convex, brown to dark brown; margin distinct and thin, about 0.1 mm wide, usually accom- panied by a white pruina. Excipulum light grey to light brown, 7-15 μm thick, internal parts appear paraplectenchymatuous and external parts prosoplec- tenchymatous. Hymenium 40-66 μm high, colourless. Hypothecium 26-74 μm high, brown to dark brown. Asci clavate, 36-48 μm×4-9 μm. Ascospores 8 per ascus, narrowly clavate, and tapering towards pro- ximal end, (3-)7-septate, 26-36 μm×2-3 μm, about 10-13 times as long as broad; I+ violet. Pycnidia not observed.
Chemistry: Trace amounts of substances were detected by TLC. Spot tests on thallus: K+ dark brown, C-, P-, KC+ dark brown.
Ecology and distribution: The species is a typi- cally foliicolous lichen, grows mainly in tropical Asia and also reported in Papua New Guinea, Australia and New Caledonia[26]. New to China.
Specimen examined: CHINA. Hainan: Wuzhishan City, Wuzhishan Nature Reserve, on leaves, 18°54′27″ N, 109°40′48″ E, elev. 730 m, 12 Dec. 2019, Y.H. Ju HN19446-a (GB accession No.: MW045559), HN19458 (GB accession No.: MW045560), HN19459 (LCUF). Changjiang County, Bawangling Nature Reserve, Bai- shitan Scenic Area, on leaves, 19°7′17″ N, 109°4′53″ E, elev. 700 m, 4 Sep, 2017, W. C. Wang HN20170025 (HMAS-L 139457, GB accession No.: MW553282); Yajia Scenic Area, on leaves, 19°7′17″N, 109°4′53″, elev. 550 m, 5 Sep. 2017. W. C. Wang HN20170124 (HMAS-L 139601, GB accession No.: MW553281).
3 DiscussionPhylogenetically Fellhanera mastothallina is close related to the type species F. fuscatula, this result confirms the hypothesis that F. mastothallina belonged to F. fuscatula group by Lücking based on evident from thallus and apothecial characters[6]. Fellhanera fuscatula can be distinguished by reddish brown apothecia, paraplectenchymatous exciple and shorter ascospores (18-24 μm×3-4.5 μm). The mor- phological characteristics of our specimens collected from Hainan are almost identical with the type speci- men from Phillippines and materials from Papua New Guinea and New Caledonia except for the longer and narrower ascospores (the latter are 22-32 μm×3-4 μm)[26-27]. Our phylogenetic result that Fellhanera being heterogeneous in its current circumscription is coincident with the preliminary studies of the family Pilocarpaceae[13, 28-29]. Some taxa within Fellhanera can be placed in other genera once sufficient evidence obtained in the future molecular studies with larger taxon sampling. Comparisons of the characteristics of the known Chinese species of Fellhanera are shown in Table 1.
| Table 1 Comparisons of the characteristics of the known Chinese species of Fellhanera |
Key to the known Chinese species of Fellhanera
1a. Lichen compounds present, mainly usnic, isousnic and zeorin acids············································································· 2
1b. Lichen compounds absent········································· ···················································································· 3
2a. Ascospores 1-septate, 10-17 μm×3-6 μm, 2.5-3.5 times as long as broad······················································· F. bouteillei
2b. Ascospores 3(-4)-septate, 10-16 μm×3-4.5 μm, 3-4 times as long as broad················································· F. subternella
3a. Ascospores l-septate or non-septate······························ ···················································································· 4
3b. Ascospores more than 1-septate·································· ···················································································· 5
4a. Ascospores oblong-ovoid, 1-septate, with constriction at septa, 10-16 μm×4-5 μm, 2.5-3.5 times as long as broad····· F. semecarpi
4b. Ascospores elongate ellipsoid, (0-)l-septate, without constriction at septa, (12-)14-17 μm×3-5.5 μm, 3-4 times as
long as broad····························································································································· F. viridisoredi
5a. Ascospores only 3-septate········································· ···················································································· 6
5b. Ascospores 5 or 7-septate, rarely 3-septate····················· ·················································································· 7
6a. Disc orange; ascospores 11-16 μm×2.5-4.5 μm, 3.5-4.5 times as long as broad················································ F. subtilis
6b. Disc reddish brown to dark greyish brown; ascospores 12-18 μm×3-4 μm, 4-5 times as long as broad··········· F. rhaphidophylli
7a. Ascospores 5-septate, rarely 3-septate··························· ·················································································· 8
7b. Ascospores 7-septate, rarely 3 or 5-septate····················· ·················································································· 9
8a. Ascospores ellipsoid, usually slightly curved and attenuated at one end, 14-21 μm×3.5-5 μm, 3.5-4.5 times as long as broad
············································································································································ F. microdiscus
8b. Ascospores oblong, with slight constrictions at septa, 14-26 μm×3-5 μm, 4-6 times as long as broad················ F. subfuscatula
9a. Ascospores oblong, 7-septate, with constrictions at septa, 18-24 μm×3-4.5 μm, 5-6 times as long as broad············· F. fuscatula
9b. Ascospores clavate, (3-)7-septate, rarely 3 or 5-septate, without constriction at septa, 26-36 μm×2-3 μm, 10-13 times
as long as broad····················································································································· F. mastothallina
| [1] |
VěZDA A. Neue gattungen der familie Lecideaceae s. lat. (Lichenes)[J]. Folia Geobot Phytotax, Praha, 1986, 21(2): 199-219. DOI:10.1007/BF02854668 |
| [2] |
LÜCKING R, HODLKINSON B P, LEAVITT S D. The 2016 classi- fication of lichenized fungi in the Ascomycota and Basidiomycota- approaching one thousand genera[J]. Bryologist, 2017, 119(4): 361-416. DOI:10.1639/0007-2745-119.4.361 |
| [3] |
KONDRATYUK S Y, LŐKÖS L, TSCHABANENKO S, et al. New and noteworthy lichen-forming and lichenicolous fungi[J]. Acta Bot Hung, 2013, 55(3/4): 275-349. DOI:10.1556/abot.55.2013.3-4.9 |
| [4] |
WIJAYAWARDENE N N, HYDE K D, KUNHIRAMAN C R, et al. Notes for genera: Ascomycota[J]. Fung Diver, 2017, 86(1): 1-594. DOI:10.1007/s13225-017-0386-0 |
| [5] |
ACHARIUS E. Förteckning pa de i Sverige växande arter af Lafvarnes famille 4[J]. Kongl Vetensk Acad Nya Handl Ser. 2, 1809, 30(3): 145-169. |
| [6] |
LÜCKING R. Foliicolous lichenized fungi[J]. Flora Neotrop, 2008, 103: 1-866. |
| [7] |
THROWER S L. Hong Kong Lichens[M]. Hong Kong: The Urban Council, 1988: 1-193.
|
| [8] |
WEI J C, JIANG Y M. Some foliicolous lichens in Xishuangbanna, China [C]//GALLOWAY D. Systematics, Conservation and Ecology of Tropical Lichens. Systematics Association Special Volume 42. Oxford: Clarendon Press, 1991: 201-216.
|
| [9] |
APTROOT A, SEAWARD M R D. Annotated checklist of Hongkong lichens[J]. Trop Bryol, 1999, 17(1): 57-101. DOI:10.11646/bde.17.1.12 |
| [10] |
APTROOT A, SIPMAN H J M. New Hong Kong lichens, ascomycetes and lichenicolous fungi[J]. J Hatt Bot Lab, 2001, 91: 317-343. |
| [11] |
APTROOT A, SPARRIUS L B. New microlichens from Taiwan[J]. Fung Diver, 2003, 14: 1-50. |
| [12] |
APTROOT A, FERRARO L I, SIMPMAN H J M, et al. Foliicolous lichens and their lichenicolous ascomycetes from Yunnan and Taiwan[J]. Mycotaxon, 2003, 88: 41-47. |
| [13] |
WANG W C, SANGVICHIEN E, WEI T Z, et al. A molecular phylo- geny of Pilocarpaceae Zahlbr., including a new species of Tapellaria Müll. Arg. and new records of foliicolous lichenized fungi from Thai- land[J]. Lichenologist, 2020, 52(5): 377-385. DOI:10.1017/S0024282920000328 |
| [14] |
CULBERSON C F, KRISTINSSON H. A standardized method for the identification of lichen products[J]. J Chromatogr, 1970, 46: 85-93. DOI:10.1016/S0021-9673(00)83967-9 |
| [15] |
CULBERSON C F. Improved conditions and new data for the identi- fication of lichen products by a standardized thin-layer chromate- graphic method[J]. J Chromatogr, 1972, 72(1): 113-125. DOI:10.1016/0021-9673(72)80013-X |
| [16] |
JIA Z F, WEI J C. Flora Lichenum Sinicorum, Vol.13, Ostropales (I), Graphidaceae 1[J]. Beijing: Science Press, 2016, 1-210. (in Chinese). |
| [17] |
ZOLLER S, SCHEIDEGGER C, SPERISEN C. PCR primers for the amplification of mitochondrial small subunit ribosomal DNA of lichen- forming ascomycetes[J]. Lichenologist, 1999, 31(5): 511-516. DOI:10.1006/lich.1999.0220 |
| [18] |
EKMAN S, ANDERSEN H L, WEDIN M. The Limitations of ancestral state reconstruction and the evolution of the ascus in the Lecanorales (Lichenized Ascomycota)[J]. Syst Biol, 2008, 57(1): 141-156. DOI:10.1080/10635150801910451 |
| [19] |
KATOH K, STANDLEY D M. MAFFT multiple sequence alignment software version 7: Improvements in performance and usability[J]. Mol Biol Evol, 2013, 30(4): 772-780. DOI:10.1093/molbev/mst010 |
| [20] |
CASTRESANA J. Selection of conserved blocks from multiple align- ments for their use in phylogenetic analysis[J]. Mol Biol Evol, 2000, 17(4): 540-552. DOI:10.1093/oxfordjournals.molbev.a026334 |
| [21] |
MILLER M A, PFEIFFER W, SCHWARTZ T. Creating the CIPRES Science Gateway for inference of large phylogenetic trees[C]//Proceedings of the Gateway Computing Environments Workshop (Gce). LA: New Orleans, 2010: 1-8.
|
| [22] |
STAMATAKIS A. RAxML Version 8: A tool for phylogenetic analysis and post-analysis of large phylogenies[J]. Bioinformatics, 2014, 30(9): 1312-1313. DOI:10.1093/bioinformatics/btu033 |
| [23] |
DARRIBA D, TABOADA G L, DOALLO R, et al. jModelTest 2: More models, new heuristics and parallel computing[J]. Nat Methods, 2012, 9: 772. DOI:10.1038/nmeth.2109 |
| [24] |
RONQUIST F, HUELSENBECK J P. MrBayes 3: Bayesian phylogenetic inference under mixed models[J]. Bioinformatics, 2003, 19: 1572-1574. DOI:10.1093/bioinformatics/btg180 |
| [25] |
RAMBAUT A. FigTree 1.2.2. [OL]. 2009. (2018-01-03) http://tree.bio.ed.ac.uk/software/figtree/.
|
| [26] |
LÜCKING R, CÁCERES M E S, KALB K, et al. Studies in Bacidia sensu lato (lichenized Ascomycetes: Lecanorales): Ⅱ. Six new combi- nations in Fellhanera Vězda[J]. Lichenologist, 2001, 33(3): 189-194. DOI:10.1006/lich.2000.0318 |
| [27] |
SANTESSON R. Foliicolous lichens: I. A revision of the taxonomy of the obligately foliicolous, lichenized fungi[J]. Symb Bot Upsal, 1952, 12(1): 1-590. |
| [28] |
ANDERSEN H L, EKMMAN S. Disintegration of the Micareaceae (lichenized Ascomycota): A molecular phylogeny based on mitochon- drial rDNA sequences[J]. Mycol Res, 2005, 109(1): 21-30. DOI:10.1017/S0953756204001625 |
| [29] |
APRTOOT A, SPARRIUS L B, ALVARADO P. Aquacidia, a new genus to accommodate a group of skiophilous temperate Bacidia species that belong in the Pilocarpaceae (lichenized ascomycetes)[J]. Gorteria, 2018, 40: 11-14. |
| [30] |
SPIRE L, APTROOT A, HERK K V. Asemone, an additional secon- dary substance in Fellhanera bouteillei in Europe[J]. Lichenologist, 2002, 34(5): 447-449. DOI:10.1006/lich.2002.0411 |
 2022, Vol. 30
2022, Vol. 30


