Scleropyrum wallichianum (Wight et Arn.) Arn. is an evergreen shrub or small trees, distributed in Guangxi, Hainan and Yunnan Provinces, China[1]. It is a potential commercial resource for extracting natural acetylenic acid, because octadec-9-ynoic was discovered in Santalaceae plants[2] and 17-octadecen-9-ynoic acid was separated from S. wallichianum[3]. Moreover, the nutlet of S. wallichianum is eaten by national minorities such as Jino nationality in Xishuangbanna[4].
According to the investigation of S. wallichianum in Xishuangbanna and Pu'er of Yunnan Province, its habitats were serious destroyed especially around the villages, urging the protection of the germplasm resources. Furthermore, as a potential commercial species, it is necessary to study on germplasm resources of S. wallichianum. There were abundant variations in the content of octadecenoic acid among different populations[5], and the phenotypic variation of S. wallichianum seeds was 93.72% within population, 6.28% among populations[6]. In addition, the content of fatty acid extracted from different varieties of S. wallichianum seeds varied greatly[7]. However, these studies were based on the morphology and component content, no molecular research has been conducted. In order to protect more genetic resources, it is necessary analysis the genetic information in S. wallichianum.
Amplified fragment length polymorphism (AFLP) markers have been widely used to investigate the genetic diversity and relationships of plant germplasm resources[8-12]. AFLP markers have a number of attractive features, the application does not require knowledge of the nucleotide sequences, highly polymorphic and readily reproducible, and the PCR-based AFLP procedure is technically simple. In addition, because AFLPs are dominant molecular markers, data inter-predation is relatively simple. In this study, the genetic diversity and relationships of 86 germplasms of S. wallichianum from seven natural populations in Yunnan Province were analyzed by using AFLP markers, which would provide a basis for introduction and conversation of S. wallichianum.
1 Materials and methods 1.1 MaterialsThe young leaves and barks were collected from 86 individuals of seven populations throughout the distribution range[13] of Scleropyrum wallichianum (Fig. 1, Table 1). The materials were dried and saved by silica gel.
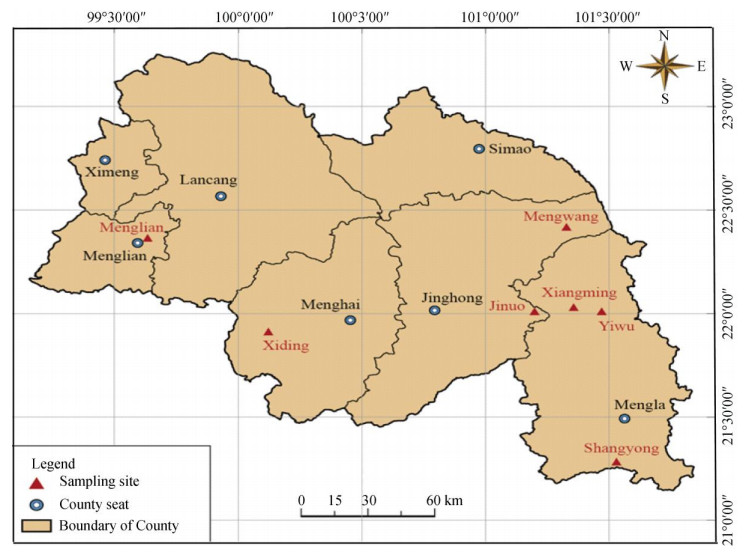
|
Fig. 1 Distribution of seven Scleropyrum wallichianum populations |
| Table 1 Geographic information of Scleropyrum wallichianum |
Genomic DNA was extracted from dried leaves or barks by using modified cetyltrimethylammonium bromide method[14]. AFLP was performed by Beijing Dingguo Changsheng Biotechnology Co. Ltd. The genomic DNA was digested with EcoR Ⅰ and Mse Ⅰ restriction endonucleases.
From 64 pairs of AFLP primer, 8 pairs with high and stable polymorphism were selected, such as EGAA/M-CAG, E-GAA/M-CTC, E-GAG/M-CAA, EGAG/M-CAC, E-GAG/M-CAG, E-GAT/M-CAA, EGAT/M-CAT and E-GAT/M-CTC, where E and M represent 5′-GACTGCGTACCAATTCA-3′ and 5′-GACGATGAGTCCTGAG-3′, respectively.
1.3 Data analysisAFLP data generated using an ABI PRISM 377 sequencer were scored as "1" (presence of fragment) and "0" (absence of fragment) by GENESCAN. The binary data matrix was first analyzed using POPGENE version 1.31 under the assumption that the populations were in Hardy-Weinberg equilibrium. The parameters of genetic diversity included number of different alleles (Na), number of effective alleles (Ne), Shannon's information index (I), Nei's gene diversity (H), the total gene diversity (HT), gene diversity within populations (HS), coefficient of genetic variation among populations (GST), gene flow (Nm), Nei's genetic distance (D) between populations. To test the genetic differentiation among populations, analysis of molecular variance (AMOVA) was conducted using the Arlequin version 3.11[15]. The longitude and latitude were converted to geographical distance data using the longitude and latitude conversion software and the Mantel test was used to test the possible association between pairwise geographical distance and genetic distance among seven populations using TFPGA version 1.3[16].
2 Results 2.1 Genetic diversityOut of 64 tested EcoR Ⅰ/Mse Ⅰ primer pairs, 8 pairs of primer showing high polymorphism were selected to evaluate and characterize the 86 germplasms of S. wallichianum. A total of 1 728 distinct bands were amplified, of which 1 388 bands were polymorphic, accounting for 80.14%. For each pair of primer, the number of amplified bands was 216, thus, the percentage of polymorphic bands amplified by AFLP primers ranged from 76.34% (E-GAA/M-CTC) to 84.88% (E-GAT/M-CAT). At the species level, the number of different alleles (Na), number of effective alleles (Ne), Nei's gene diversity (H) and Shannon's information index (I) were 1.417, 1.179, 0.137, and 0.225, respectively. At the population level, Nei's gene diversity (H) and the Shannon's information index (I) were estimated to be 0.111 and 0.175, respectively.
2.2 Genetic structureWhen all the seven populations were considered, the coefficient of genetic variation among populations (GST) was 0.191. The total gene diversity (HT) and gene diversity within populations (HS) were 0.138 and 0.111, respectively. AMOVA showed that 22.7% of the total genetic variation occurred among populations and 78.3% within each population with average pairwise φPT (similar to FST) of 0.135. The estimated gene flow (Nm) among populations was 2.141.
2.3 Genetic relationshipsAt a genetic similarity coefficient of 0.52, all the germplasms were classified into three clusters in UPGMA tree, such as cluster Ⅰ, Ⅱ and Ⅲ (Fig. 2). Cluster Ⅰ was comprised of 47 germplasms, accounting for 54.65%, while cluster Ⅱ and cluster Ⅲ with 19 and 20 germplasms, accounting for 22.09% and 23.26%, respectively. Cluster Ⅰ could be further divided into Ia and Ib groups with 24 and 23 germplasms, respectively.
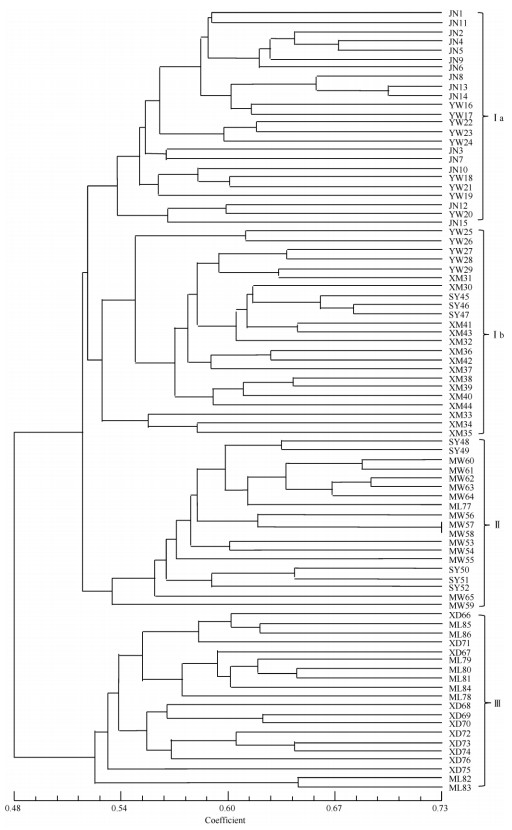
|
Fig. 2 UPGMA tree of Scleropyrum wallichianum germplasms |
Among populations, Nei's unbiased genetic identity (In) ranged from 0.9873 (between YW and XM population) to 0.936 (between JN and XD population), with an average of 0.977. The smallest genetic distance (0.015) was detected between population YW and XM, the biggest genetic distance (0.073) was detected between population SY and XM, as well as population XD and JN (0.072) (Table 2). The relationships among seven populations were analyzed based on Nei's unbiased genetic identity (In) by using UPGMA clustering method (Fig. 3). Population YW and XM were grouped into a cluster at first, following population SY and MW, then with population ML and XD again, and finally with population JN. There was no significant correlation between genetic distance and geographical distance among all populations (r=0.0323, P=0.5820) by Mantel test.
| Table 2 Nei's unbiased genetic identity and genetic distance among populations |
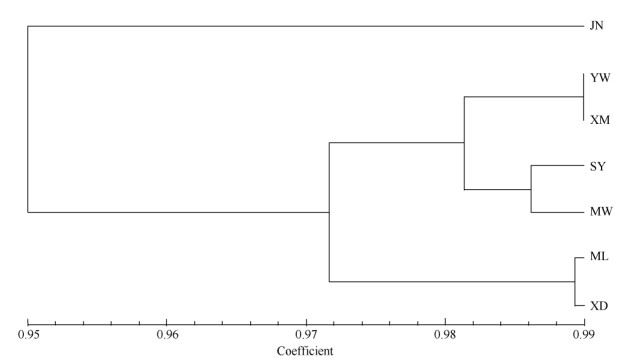
|
Fig. 3 UPGMA tree of Scleropyrum wallichianum populations |
According to phenotype data of seeds, Wu et al.[6]reported that there were big phenotype variations (93.72%) within populations of S. wallichianum, and small variations among seven populations (6.28%). Based on the content of fatty acid of S. wallichianum, Wu et al.[7] considered that the variation within populations was larger than that among populations. In our study, similar results was obtained on the basis of the AFLP data, which revealed that 78.3% of the total variations distributed within populations, and 22.7% among populations.
The high genetic diversity of S. wallichianum may be attributed to its biological characteristics and geographical distribution. According to the record of Floral of China, Scleropyrum wallichianum mainly distributed in Guangxi, Hainan, Yunnan Provinces, and Indo-China Peninsula. Taking our sampling into consideration, these samples gathered in Yunnan represented the northern border of distribution area of the species, where historically complex tectonic events continually taken place. It is possible that these samples had sympatric distribution areas before the Yunnan-Guizhou Plateau uplift, causing low level of genetic differentiation between populations. In addition, the latitudes of S. wallichianum distributed range from 900~1 600 m, and grow in mixed forest habitats. Long-term adaptability to diverse environments was conducive to the accumulation and preservation of genetic variation. Even single and small distribution area was formed after geological uplifting, but the genetic variation speed among population was slower than that of within populations.
3.2 Genetic structureIn this study, a high level of genetic differentiation among population was detected with GST=0.191, and φPT=0.135. Among the many factors influencing interpopulation genetic differentiations, gene flow was the most obvious. Population experiencing high gene flow tended to have lower genetic differentiation than that of populations with limited gene flow. In this study, Nm was estimated to be 2.141, which suggested the existence of a strong gene exchange among these seven populations. The proliferation and migration of individuals or propagules was associated with gene flow among different populations[17]. According to our observation in the wild field, Scleropyrum wallichianum relies mainly on animals for seed dispersal, which provided opportunities for genetic recombination between populations. For pollen dispersal, because no investigation and studies recorded for this species, we don't know whether or not the pollen dispersal increase the gene flow between populations.
3.3 Genetic relationshipsAccording to the results of cluster analysis based on genetic similarity, most germplasms from the same population mixed with other clusters (Fig. 2), which suggested wide genetic variability existed in these seven populations. The habitat of all populations was hillside and ridge. Similar environment may be reasonable for the cross-distribution of these germplasms. At population level, population YW and XM from Mengla County was clustered at first, they and populations SY and MW then clustered into one group, which had close genetic relationship with the cluster of populations ML and XD. Population JN, however, was only distantly related to the other populations. Population ML and SY had the farthest geographical distance (219.1 km) (Table 2 and Fig. 3), but their genetic distance was not the largest. This suggested that genetic relationship was not strictly related to geographic distance among these seven populations. Furthermore, there was no significant correlation between genetic distance and geographical distance (r=0.0323, P=0.5820) for these populations as a whole.
4 Introduction and conservation strategyAs a potential resource for extracting natural acetylenic acid, and the material of nut foods, Scleropyrum wallichianum should be paid more attention, especially to the natural resource exploring and conservation. Therefore, our results in this paper could provide necessary information for selecting excellent individuals and germplasm resources with abundant variation. Eleven germplasms, such as JN14, MW63, JN13, YW18, XD75, YW25, MW62, YW17, SY49, JN8 and ML78, were selected with high polymorphism of specific loci. These germplasms, accounting for 20.53% of total specific bands, should be priority in protection and attention.
For commercial resource, the seeds of S. wallichianum were valuable, so, the fruitful germplasms should be introduced. But for species diversity, the germplasms with large genetic variation should be protected. From this study and based on the present situation of S. wallichianum distribution, we consequently proposed the following conservation strategies. Partly germplasms can be protected in situ, because this species mainly grow around the village, accompanying with farming plant, and the seeds can provide a fine food for local people. For ex situ conservation, the eleven germplasms above-mentioned can be introduced in order to increase genetic diversity. Additionally, these eleven germplasms should be well maintain the sustainable development of existing genetic diversity of S. wallichianum.
Acknowledgment We thank Dr. Zheng Ying for revising the article.
| [1] |
WU Z Y, RAVEN P H, HONG D Y. Flora of China, Vol. 5[M]. Beijing: Science Press & St. Louis: Missouri Botanical Garden Press, 2003: 208-219.
|
| [2] |
Morris L J, Marshall M O. Occurrence of stearolic acid in Santalaceae seed oils[J]. Chem Ind, 1966, 12: 460-461. |
| [3] |
Wang J P, LI J M, Yu F L. Acetylenic acids from seed oil of Scleropyrum wallichianum[J]. Acta Bot Yunnan, 1992, 14(1): 101-104. (in Chinese). |
| [4] |
Wang J R, Long C L. Ethnobotanical study of traditional edible plants of Jinuo nationality[J]. Acta Bot Yunnan, 1995, 17(2): 161-168. (in Chinese). |
| [5] |
Editorial Committee of Oil Plants in China. Oil Plants in China[M]. Beijing: Science Press, 1987: 90-92. (in Chinese).
|
| [6] |
Wu Y, He M Y, Zhang F L, et al. Phenotype variations of seed in wilding Scleropyrum wallichianum[J]. J NE For Univ, 2015, 43(8): 29-33. (in Chinese). |
| [7] |
WU Y, HE M Y, ZHANG F L, et al. Population variation of fatty acids from the seeds wild oil-bearing tree Scleropyrum wallichianum in Yunnan[J]. J NE For Univ, 2015, 30(6): 99-103. (in Chinese). DOI:10.3969/j.issn.1001-7461.2015.06.17 |
| [8] |
QIN X B, GAO J H. Studies on genetic diversity for specialty kiwifruits from southwestern regions of China using AFLP system[J]. J Trop Subtrop Bot, 2013, 21(4): 315-322. (in Chinese). DOI:10.3969/j.issn.1005-3395.2013.04.004 |
| [9] |
ZHANG Y, RAN H, FENG Y, et al. Studies on the relationship of newly published infra-species of Phyllostachys edulis by AFLP marker[J]. J Trop Subtrop Bot, 2016, 24(6): 657-664. (in Chinese). DOI:10.11926/j.issn.1005-3395.2016.06.009 |
| [10] |
Rapposelli E, Melito S, Barmina G G, et al. AFLP finger printing and essential oil profiling of cultivated and wild populations of Sardinian Salvia desoleana[J]. Genet Resour Crop Evol, 2015, 62(6): 959-970. DOI:10.1007/s10722-015-0263-1 |
| [11] |
Liu J Y, Zhao P F, Liu X L, et al. Genetic diversity analysis on 68 foreign sugarcane germplasms (Saccharum spp.) with AFLP technique[J]. J Hunan Agric Univ, 2013, 39(5): 466-470. (in Chinese). |
| [12] |
WANG X, Xing S Y, Sun L M, et al. Genetic diversity of Styrax obassia Sieb. et Zucc. based on AFLP markers[J]. Biochem Syst Ecol, 2015, 61: 28-34. DOI:10.1016/j.bse.2015.04.024 |
| [13] |
WU Y, Zhang F L, He M Y, et al. Investigation of wild resource of oil tree Scleropyrum wallichianum in Yunnan[J]. Trop Agric Sci Technol, 2015, 38(1): 24-26. (in Chinese). DOI:10.3969/j.issn.1672-450X.2015.01.007 |
| [14] |
Doyle J. DNA protocols for plants [M]// Hewitt G M, Johnston A W B, YOUNG J P W. Molecular Techniques in Taxonomy. Berlin: Springer, 1991: 283-293. doi: 10.1007/978-3-642-83962-7_18.
|
| [15] |
Excoffier L, Laval G, Schneider S. Arlequin (version 3.0): An integrated software package for population genetics data analysis[J]. Evol Bioinform, 2005, 1: 47-50. DOI:10.1177/117693430500100003 |
| [16] |
Miller M P. Tools for Population Genetic Analyses (TFPGA), Ver. 1.3 A [CP/OL]. [2018-01-10] http://www.marksgeneticsoftware.net/_vti_bin/shtml.exe/tfpga.htm?
|
| [17] |
Levin D A, Kerster H W. Gene flow in seed plants [M]// DOBZHANSKY T, HECHT M K, STEERE W C. Evolutionary Biology. Boston, MA: Springer, 1974: 139-220. doi: 10.1007/978-1-4615-944-2_5.
|
 2019, Vol. 27
2019, Vol. 27


